Devrim Unay
COVID-19 Detection Using Transfer Learning Approach from Computed Tomography Images
Jul 08, 2022



Abstract:Our main goal in this study is to propose a transfer learning based method for COVID-19 detection from Computed Tomography (CT) images. The transfer learning model used for the task is a pretrained Xception model. Both model architecture and pre-trained weights on ImageNet were used. The resulting modified model was trained with 128 batch size and 224x224, 3 channeled input images, converted from original 512x512, grayscale images. The dataset used is a the COV19-CT-DB. Labels in the dataset include COVID-19 cases and Non-COVID-19 cases for COVID-1919 detection. Firstly, a accuracy and loss on the validation partition of the dataset as well as precision recall and macro F1 score were used to measure the performance of the proposed method. The resulting Macro F1 score on the validation set exceeded the baseline model.
Deep Learning Based Automated COVID-19 Classification from Computed Tomography Images
Nov 22, 2021

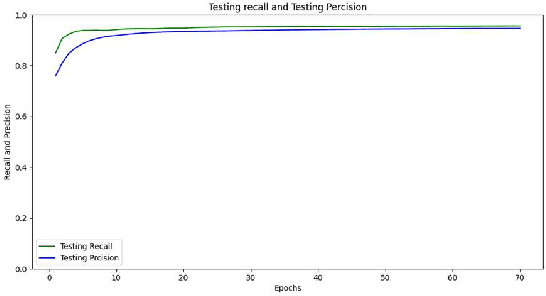

Abstract:The paper presents a Convolutional Neural Networks (CNN) model for image classification, aiming at increasing predictive performance for COVID-19 diagnosis while avoiding deeper and thus more complex alternatives. The proposed model includes four similar convolutional layers followed by a flattening and two dense layers. This work proposes a less complex solution based on simply classifying 2D CT-Scan slices of images using their pixels via a 2D CNN model. Despite the simplicity in architecture, the proposed model showed improved quantitative results exceeding state-of-the-art on the same dataset of images, in terms of the macro f1 score. In this case study, extracting features from images, segmenting parts of the images, or other more complex techniques, ultimately aiming at images classification, do not yield better results. With that, this paper introduces a simple yet powerful deep learning based solution for automated COVID-19 classification.
Channel Attention Networks for Robust MR Fingerprinting Matching
Dec 02, 2020



Abstract:Magnetic Resonance Fingerprinting (MRF) enables simultaneous mapping of multiple tissue parameters such as T1 and T2 relaxation times. The working principle of MRF relies on varying acquisition parameters pseudo-randomly, so that each tissue generates its unique signal evolution during scanning. Even though MRF provides faster scanning, it has disadvantages such as erroneous and slow generation of the corresponding parametric maps, which needs to be improved. Moreover, there is a need for explainable architectures for understanding the guiding signals to generate accurate parametric maps. In this paper, we addressed both of these shortcomings by proposing a novel neural network architecture consisting of a channel-wise attention module and a fully convolutional network. The proposed approach, evaluated over 3 simulated MRF signals, reduces error in the reconstruction of tissue parameters by 8.88% for T1 and 75.44% for T2 with respect to state-of-the-art methods. Another contribution of this study is a new channel selection method: attention-based channel selection. Furthermore, the effect of patch size and temporal frames of MRF signal on channel reduction are analyzed by employing a channel-wise attention.
Combining nonparametric spatial context priors with nonparametric shape priors for dendritic spine segmentation in 2-phoyon microscopy images
Jan 08, 2019


Abstract:Data driven segmentation is an important initial step of shape prior-based segmentation methods since it is assumed that the data term brings a curve to a plausible level so that shape and data terms can then work together to produce better segmentations. When purely data driven segmentation produces poor results, the final segmentation is generally affected adversely. One challenge faced by many existing data terms is due to the fact that they consider only pixel intensities to decide whether to assign a pixel to the foreground or to the background region. When the distributions of the foreground and background pixel intensities have significant overlap, such data terms become ineffective, as they produce uncertain results for many pixels in a test image. In such cases, using prior information about the spatial context of the object to be segmented together with the data term can bring a curve to a plausible stage, which would then serve as a good initial point to launch shape-based segmentation. In this paper, we propose a new segmentation approach that combines nonparametric context priors with a learned-intensity-based data term and nonparametric shape priors. We perform experiments for dendritic spine segmentation in both 2D and 3D 2-photon microscopy images. The experimental results demonstrate that using spatial context priors leads to significant improvements.
Dendritic Spine Shape Analysis: A Clustering Perspective
Jul 19, 2016

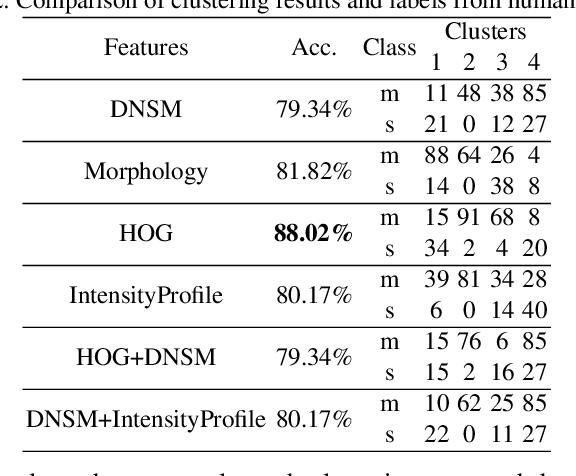

Abstract:Functional properties of neurons are strongly coupled with their morphology. Changes in neuronal activity alter morphological characteristics of dendritic spines. First step towards understanding the structure-function relationship is to group spines into main spine classes reported in the literature. Shape analysis of dendritic spines can help neuroscientists understand the underlying relationships. Due to unavailability of reliable automated tools, this analysis is currently performed manually which is a time-intensive and subjective task. Several studies on spine shape classification have been reported in the literature, however, there is an on-going debate on whether distinct spine shape classes exist or whether spines should be modeled through a continuum of shape variations. Another challenge is the subjectivity and bias that is introduced due to the supervised nature of classification approaches. In this paper, we aim to address these issues by presenting a clustering perspective. In this context, clustering may serve both confirmation of known patterns and discovery of new ones. We perform cluster analysis on two-photon microscopic images of spines using morphological, shape, and appearance based features and gain insights into the spine shape analysis problem. We use histogram of oriented gradients (HOG), disjunctive normal shape models (DNSM), morphological features, and intensity profile based features for cluster analysis. We use x-means to perform cluster analysis that selects the number of clusters automatically using the Bayesian information criterion (BIC). For all features, this analysis produces 4 clusters and we observe the formation of at least one cluster consisting of spines which are difficult to be assigned to a known class. This observation supports the argument of intermediate shape types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge