Mujdat Cetin
Conformalized Generative Bayesian Imaging: An Uncertainty Quantification Framework for Computational Imaging
Apr 10, 2025Abstract:Uncertainty quantification plays an important role in achieving trustworthy and reliable learning-based computational imaging. Recent advances in generative modeling and Bayesian neural networks have enabled the development of uncertainty-aware image reconstruction methods. Current generative model-based methods seek to quantify the inherent (aleatoric) uncertainty on the underlying image for given measurements by learning to sample from the posterior distribution of the underlying image. On the other hand, Bayesian neural network-based approaches aim to quantify the model (epistemic) uncertainty on the parameters of a deep neural network-based reconstruction method by approximating the posterior distribution of those parameters. Unfortunately, an ongoing need for an inversion method that can jointly quantify complex aleatoric uncertainty and epistemic uncertainty patterns still persists. In this paper, we present a scalable framework that can quantify both aleatoric and epistemic uncertainties. The proposed framework accepts an existing generative model-based posterior sampling method as an input and introduces an epistemic uncertainty quantification capability through Bayesian neural networks with latent variables and deep ensembling. Furthermore, by leveraging the conformal prediction methodology, the proposed framework can be easily calibrated to ensure rigorous uncertainty quantification. We evaluated the proposed framework on magnetic resonance imaging, computed tomography, and image inpainting problems and showed that the epistemic and aleatoric uncertainty estimates produced by the proposed framework display the characteristic features of true epistemic and aleatoric uncertainties. Furthermore, our results demonstrated that the use of conformal prediction on top of the proposed framework enables marginal coverage guarantees consistent with frequentist principles.
LG-Sleep: Local and Global Temporal Dependencies for Mice Sleep Scoring
Dec 19, 2024



Abstract:Efficiently identifying sleep stages is crucial for unraveling the intricacies of sleep in both preclinical and clinical research. The labor-intensive nature of manual sleep scoring, demanding substantial expertise, has prompted a surge of interest in automated alternatives. Sleep studies in mice play a significant role in understanding sleep patterns and disorders and underscore the need for robust scoring methodologies. In response, this study introduces LG-Sleep, a novel subject-independent deep neural network architecture designed for mice sleep scoring through electroencephalogram (EEG) signals. LG-Sleep extracts local and global temporal transitions within EEG signals to categorize sleep data into three stages: wake, rapid eye movement (REM) sleep, and non-rapid eye movement (NREM) sleep. The model leverages local and global temporal information by employing time-distributed convolutional neural networks to discern local temporal transitions in EEG data. Subsequently, features derived from the convolutional filters traverse long short-term memory blocks, capturing global transitions over extended periods. Crucially, the model is optimized in an autoencoder-decoder fashion, facilitating generalization across distinct subjects and adapting to limited training samples. Experimental findings demonstrate superior performance of LG-Sleep compared to conventional deep neural networks. Moreover, the model exhibits good performance across different sleep stages even when tasked with scoring based on limited training samples.
Integrating Generative and Physics-Based Models for Ptychographic Imaging with Uncertainty Quantification
Dec 14, 2024Abstract:Ptychography is a scanning coherent diffractive imaging technique that enables imaging nanometer-scale features in extended samples. One main challenge is that widely used iterative image reconstruction methods often require significant amount of overlap between adjacent scan locations, leading to large data volumes and prolonged acquisition times. To address this key limitation, this paper proposes a Bayesian inversion method for ptychography that performs effectively even with less overlap between neighboring scan locations. Furthermore, the proposed method can quantify the inherent uncertainty on the ptychographic object, which is created by the ill-posed nature of the ptychographic inverse problem. At a high level, the proposed method first utilizes a deep generative model to learn the prior distribution of the object and then generates samples from the posterior distribution of the object by using a Markov Chain Monte Carlo algorithm. Our results from simulated ptychography experiments show that the proposed framework can consistently outperform a widely used iterative reconstruction algorithm in cases of reduced overlap. Moreover, the proposed framework can provide uncertainty estimates that closely correlate with the true error, which is not available in practice. The project website is available here.
Robust EEG-based Emotion Recognition Using an Inception and Two-sided Perturbation Model
Apr 21, 2024Abstract:Automated emotion recognition using electroencephalogram (EEG) signals has gained substantial attention. Although deep learning approaches exhibit strong performance, they often suffer from vulnerabilities to various perturbations, like environmental noise and adversarial attacks. In this paper, we propose an Inception feature generator and two-sided perturbation (INC-TSP) approach to enhance emotion recognition in brain-computer interfaces. INC-TSP integrates the Inception module for EEG data analysis and employs two-sided perturbation (TSP) as a defensive mechanism against input perturbations. TSP introduces worst-case perturbations to the model's weights and inputs, reinforcing the model's elasticity against adversarial attacks. The proposed approach addresses the challenge of maintaining accurate emotion recognition in the presence of input uncertainties. We validate INC-TSP in a subject-independent three-class emotion recognition scenario, demonstrating robust performance.
Multi-Source Domain Adaptation with Transformer-based Feature Generation for Subject-Independent EEG-based Emotion Recognition
Jan 04, 2024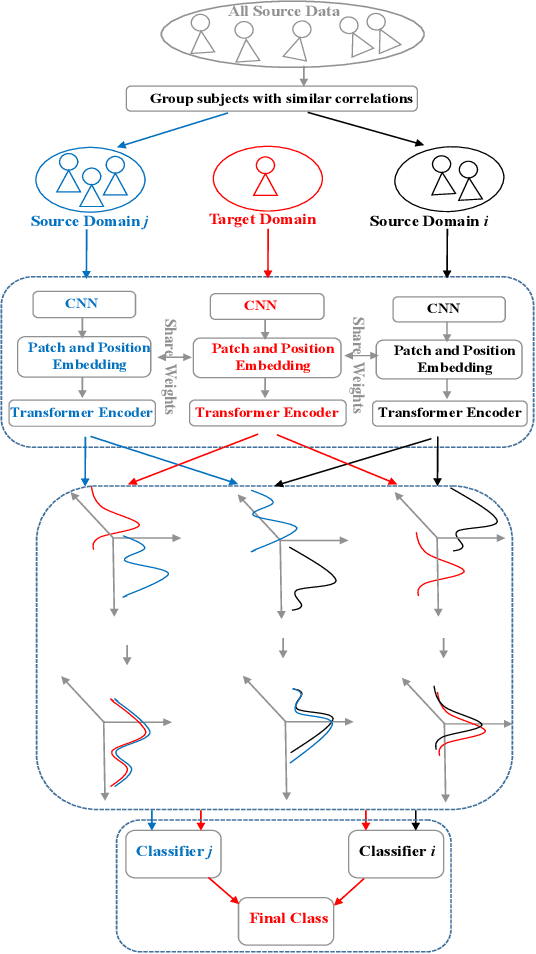
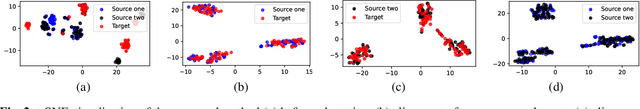
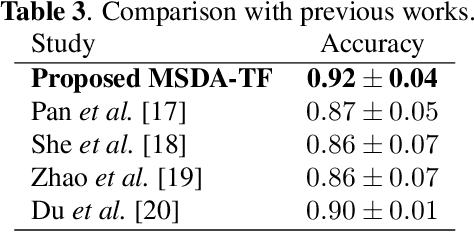
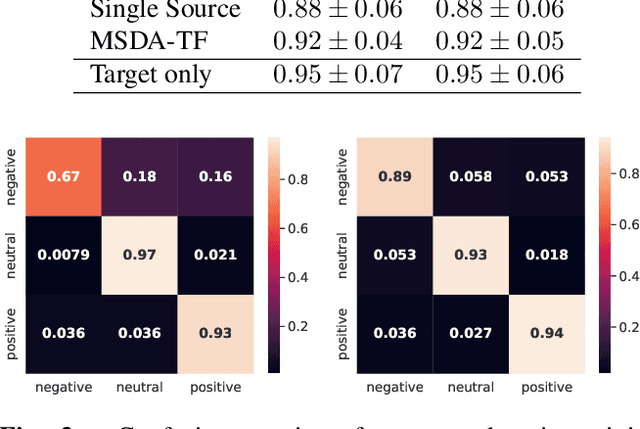
Abstract:Although deep learning-based algorithms have demonstrated excellent performance in automated emotion recognition via electroencephalogram (EEG) signals, variations across brain signal patterns of individuals can diminish the model's effectiveness when applied across different subjects. While transfer learning techniques have exhibited promising outcomes, they still encounter challenges related to inadequate feature representations and may overlook the fact that source subjects themselves can possess distinct characteristics. In this work, we propose a multi-source domain adaptation approach with a transformer-based feature generator (MSDA-TF) designed to leverage information from multiple sources. The proposed feature generator retains convolutional layers to capture shallow spatial, temporal, and spectral EEG data representations, while self-attention mechanisms extract global dependencies within these features. During the adaptation process, we group the source subjects based on correlation values and aim to align the moments of the target subject with each source as well as within the sources. MSDA-TF is validated on the SEED dataset and is shown to yield promising results.
A Hybrid End-to-End Spatio-Temporal Attention Neural Network with Graph-Smooth Signals for EEG Emotion Recognition
Jul 06, 2023Abstract:Recently, physiological data such as electroencephalography (EEG) signals have attracted significant attention in affective computing. In this context, the main goal is to design an automated model that can assess emotional states. Lately, deep neural networks have shown promising performance in emotion recognition tasks. However, designing a deep architecture that can extract practical information from raw data is still a challenge. Here, we introduce a deep neural network that acquires interpretable physiological representations by a hybrid structure of spatio-temporal encoding and recurrent attention network blocks. Furthermore, a preprocessing step is applied to the raw data using graph signal processing tools to perform graph smoothing in the spatial domain. We demonstrate that our proposed architecture exceeds state-of-the-art results for emotion classification on the publicly available DEAP dataset. To explore the generality of the learned model, we also evaluate the performance of our architecture towards transfer learning (TL) by transferring the model parameters from a specific source to other target domains. Using DEAP as the source dataset, we demonstrate the effectiveness of our model in performing cross-modality TL and improving emotion classification accuracy on DREAMER and the Emotional English Word (EEWD) datasets, which involve EEG-based emotion classification tasks with different stimuli.
Uncertainty Quantification for Deep Unrolling-Based Computational Imaging
Jul 02, 2022



Abstract:Deep unrolling is an emerging deep learning-based image reconstruction methodology that bridges the gap between model-based and purely deep learning-based image reconstruction methods. Although deep unrolling methods achieve state-of-the-art performance for imaging problems and allow the incorporation of the observation model into the reconstruction process, they do not provide any uncertainty information about the reconstructed image, which severely limits their use in practice, especially for safety-critical imaging applications. In this paper, we propose a learning-based image reconstruction framework that incorporates the observation model into the reconstruction task and that is capable of quantifying epistemic and aleatoric uncertainties, based on deep unrolling and Bayesian neural networks. We demonstrate the uncertainty characterization capability of the proposed framework on magnetic resonance imaging and computed tomography reconstruction problems. We investigate the characteristics of the epistemic and aleatoric uncertainty information provided by the proposed framework to motivate future research on utilizing uncertainty information to develop more accurate, robust, trustworthy, uncertainty-aware, learning-based image reconstruction and analysis methods for imaging problems. We show that the proposed framework can provide uncertainty information while achieving comparable reconstruction performance to state-of-the-art deep unrolling methods.
Combining physics-based modeling and deep learning for ultrasound elastography
Jul 28, 2021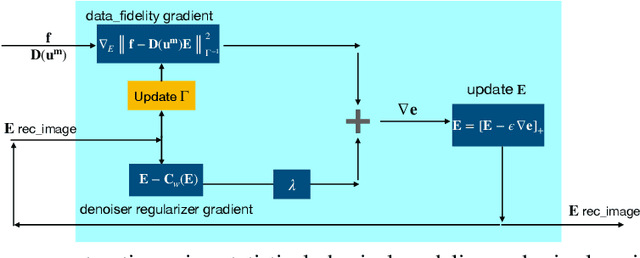

Abstract:Ultrasound elasticity images which enable the visualization of quantitative maps of tissue stiffness can be reconstructed by solving an inverse problem. Classical model-based approaches for ultrasound elastography use deterministic finite element methods (FEMs) to incorporate the governing physical laws resulting in poor performance in noisy conditions. Moreover, these approaches utilize fixed regularizers for various tissue patterns while appropriate data-adaptive priors might be required for capturing the complex spatial elasticity distribution. In this regard, we propose a joint model-based and learning-based framework for estimating the elasticity distribution by solving a regularized optimization problem. We present an integrated objective function composed of a statistical physics-based forward model and a data-driven regularizer to leverage deep neural networks for learning the underlying elasticity prior. This constrained optimization problem is solved using the gradient descent (GD) method and the gradient of regularizer is simply replaced by the residual of the trained denoiser network for having an explicit objective function with reduced computation time.
Regularization by Adversarial Learning for Ultrasound Elasticity Imaging
Jun 14, 2021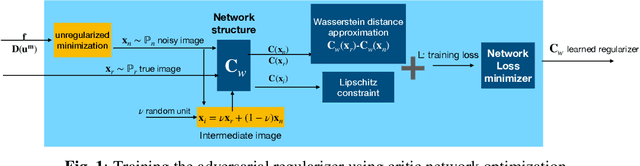
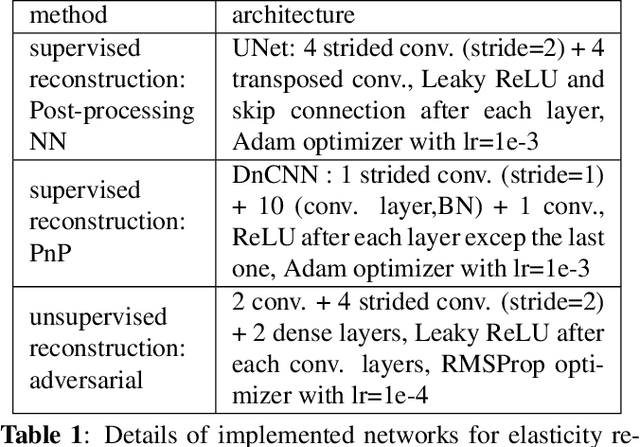
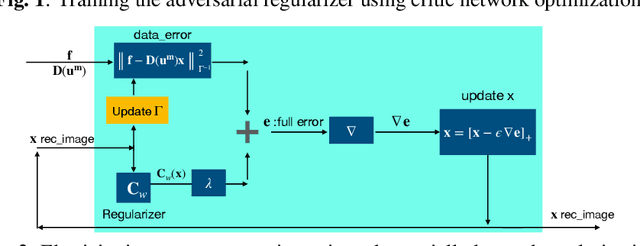
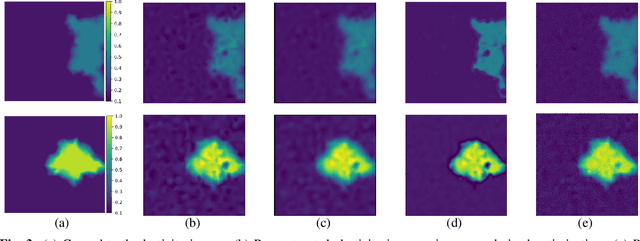
Abstract:Classical model-based imaging methods for ultrasound elasticity inverse problem require prior constraints about the underlying elasticity patterns, while finding the appropriate hand-crafted prior for each tissue type is a challenge. In contrast, standard data-driven methods count solely on supervised learning on the training data pairs leading to massive network parameters for unnecessary physical model relearning which might not be consistent with the governing physical models of the imaging system. Fusing the physical forward model and noise statistics with data-adaptive priors leads to a united reconstruction framework that guarantees the learned reconstruction agrees with the physical models while coping with the limited training data. In this paper, we propose a new methodology for estimating the elasticity image by solving a regularized optimization problem which benefits from the physics-based modeling via a data-fidelity term and adversarially learned priors via a regularization term. In this method, the regularizer is trained based on the Wasserstein Generative Adversarial Network (WGAN) objective function which tries to distinguish the distribution of clean and noisy images. Leveraging such an adversarial regularizer for parameterizing the distribution of latent images and using gradient descent (GD) for solving the corresponding regularized optimization task leads to stability and convergence of the reconstruction compared to pixel-wise supervised learning schemes. Our simulation results verify the effectiveness and robustness of the proposed methodology with limited training datasets.
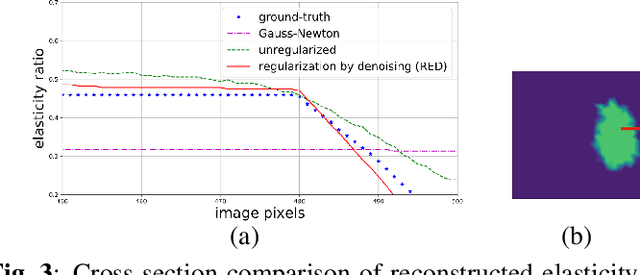
MR elasticity reconstruction using statistical physical modeling and explicit data-driven denoising regularizer
May 27, 2021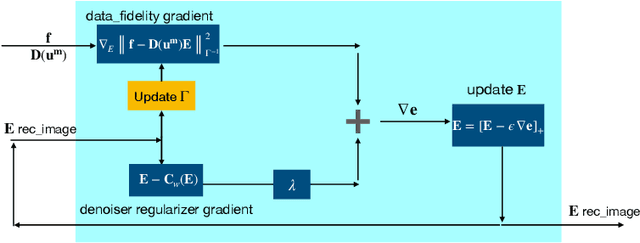
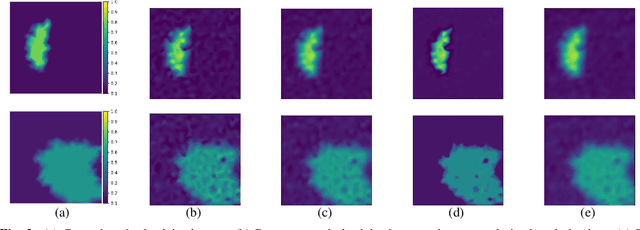
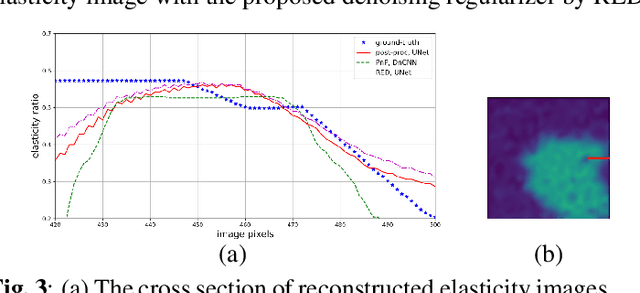
Abstract:Elasticity image, visualizing the quantitative map of tissue stiffness, can be reconstructed by solving an inverse problem. Classical methods for magnetic resonance elastography (MRE) try to solve a regularized optimization problem comprising a deterministic physical model and a prior constraint as data-fidelity term and regularization term, respectively. For improving the elasticity reconstructions, appropriate prior about the underlying elasticity distribution is required which is not unique. This article proposes an infused approach for MRE reconstruction by integrating the statistical representation of the physical laws of harmonic motions and learning-based prior. For data-fidelity term, we use a statistical linear-algebraic model of equilibrium equations and for the regularizer, data-driven regularization by denoising (RED) is utilized. In the proposed optimization paradigm, the regularizer gradient is simply replaced by the residual of learned denoiser leading to time-efficient computation and convex explicit objective function. Simulation results of elasticity reconstruction verify the effectiveness of the proposed approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge