Derek C. Rose
Hyperparameter Optimization in Binary Communication Networks for Neuromorphic Deployment
Apr 21, 2020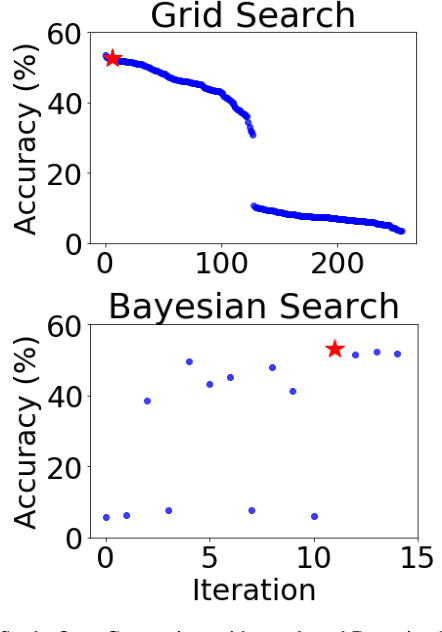
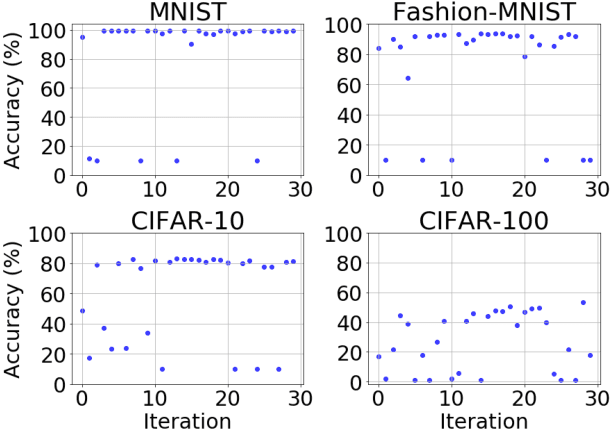
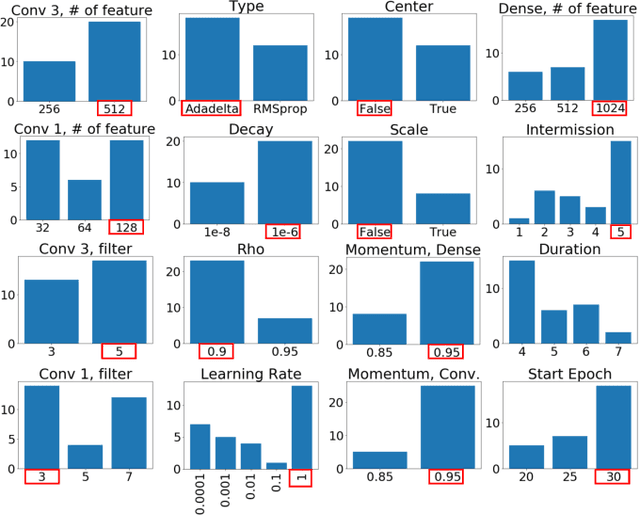
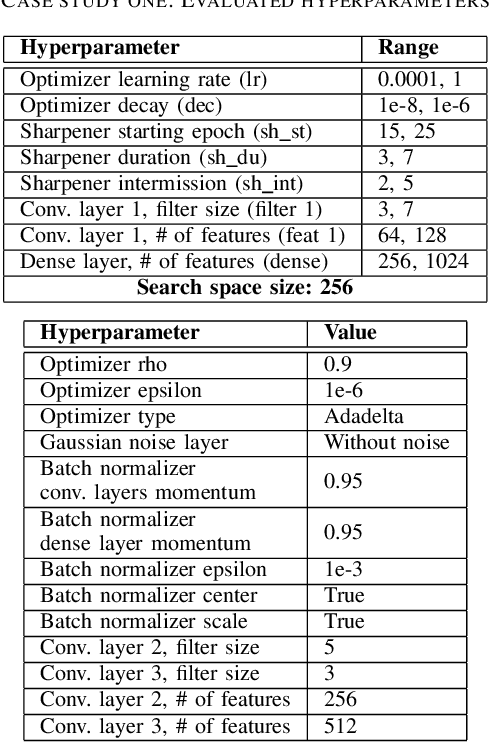
Abstract:Training neural networks for neuromorphic deployment is non-trivial. There have been a variety of approaches proposed to adapt back-propagation or back-propagation-like algorithms appropriate for training. Considering that these networks often have very different performance characteristics than traditional neural networks, it is often unclear how to set either the network topology or the hyperparameters to achieve optimal performance. In this work, we introduce a Bayesian approach for optimizing the hyperparameters of an algorithm for training binary communication networks that can be deployed to neuromorphic hardware. We show that by optimizing the hyperparameters on this algorithm for each dataset, we can achieve improvements in accuracy over the previous state-of-the-art for this algorithm on each dataset (by up to 15 percent). This jump in performance continues to emphasize the potential when converting traditional neural networks to binary communication applicable to neuromorphic hardware.
Exascale Deep Learning to Accelerate Cancer Research
Sep 26, 2019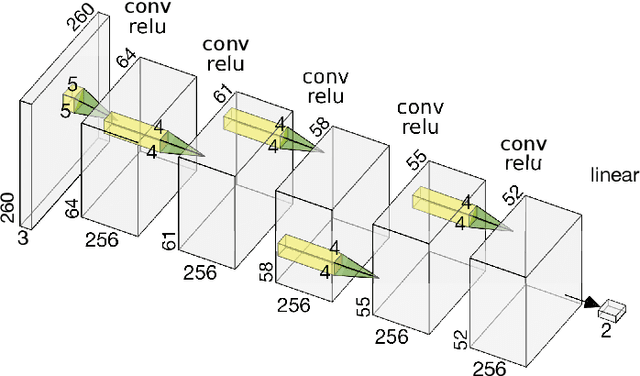
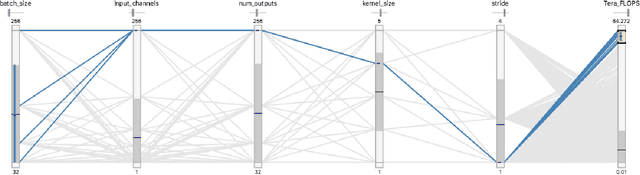
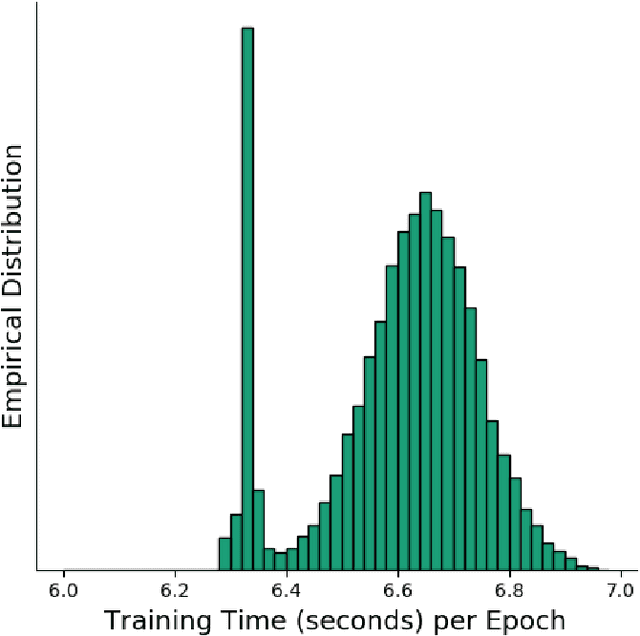
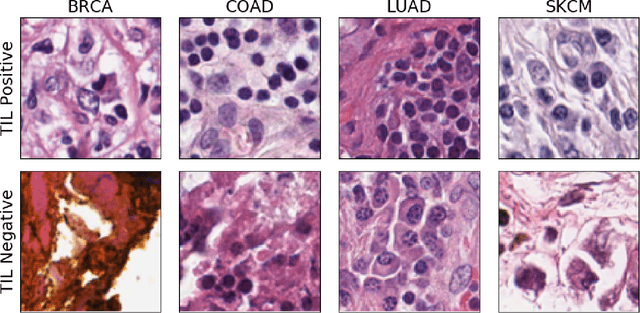
Abstract:Deep learning, through the use of neural networks, has demonstrated remarkable ability to automate many routine tasks when presented with sufficient data for training. The neural network architecture (e.g. number of layers, types of layers, connections between layers, etc.) plays a critical role in determining what, if anything, the neural network is able to learn from the training data. The trend for neural network architectures, especially those trained on ImageNet, has been to grow ever deeper and more complex. The result has been ever increasing accuracy on benchmark datasets with the cost of increased computational demands. In this paper we demonstrate that neural network architectures can be automatically generated, tailored for a specific application, with dual objectives: accuracy of prediction and speed of prediction. Using MENNDL--an HPC-enabled software stack for neural architecture search--we generate a neural network with comparable accuracy to state-of-the-art networks on a cancer pathology dataset that is also $16\times$ faster at inference. The speedup in inference is necessary because of the volume and velocity of cancer pathology data; specifically, the previous state-of-the-art networks are too slow for individual researchers without access to HPC systems to keep pace with the rate of data generation. Our new model enables researchers with modest computational resources to analyze newly generated data faster than it is collected.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge