Daniel Jeong
Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA, 91109, USA
Reliable Radiologic Skeletal Muscle Area Assessment -- A Biomarker for Cancer Cachexia Diagnosis
Mar 19, 2025Abstract:Cancer cachexia is a common metabolic disorder characterized by severe muscle atrophy which is associated with poor prognosis and quality of life. Monitoring skeletal muscle area (SMA) longitudinally through computed tomography (CT) scans, an imaging modality routinely acquired in cancer care, is an effective way to identify and track this condition. However, existing tools often lack full automation and exhibit inconsistent accuracy, limiting their potential for integration into clinical workflows. To address these challenges, we developed SMAART-AI (Skeletal Muscle Assessment-Automated and Reliable Tool-based on AI), an end-to-end automated pipeline powered by deep learning models (nnU-Net 2D) trained on mid-third lumbar level CT images with 5-fold cross-validation, ensuring generalizability and robustness. SMAART-AI incorporates an uncertainty-based mechanism to flag high-error SMA predictions for expert review, enhancing reliability. We combined the SMA, skeletal muscle index, BMI, and clinical data to train a multi-layer perceptron (MLP) model designed to predict cachexia at the time of cancer diagnosis. Tested on the gastroesophageal cancer dataset, SMAART-AI achieved a Dice score of 97.80% +/- 0.93%, with SMA estimated across all four datasets in this study at a median absolute error of 2.48% compared to manual annotations with SliceOmatic. Uncertainty metrics-variance, entropy, and coefficient of variation-strongly correlated with SMA prediction errors (0.83, 0.76, and 0.73 respectively). The MLP model predicts cachexia with 79% precision, providing clinicians with a reliable tool for early diagnosis and intervention. By combining automation, accuracy, and uncertainty awareness, SMAART-AI bridges the gap between research and clinical application, offering a transformative approach to managing cancer cachexia.
Assistive Relative Pose Estimation for On-orbit Assembly using Convolutional Neural Networks
Feb 19, 2020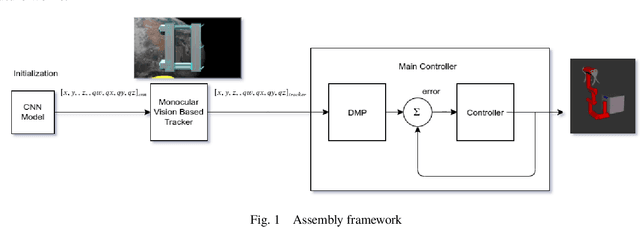
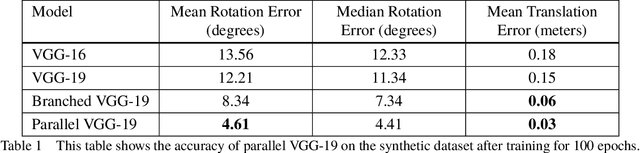
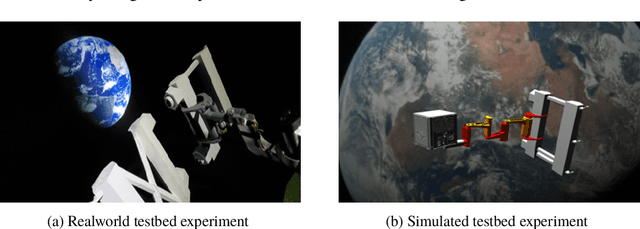
Abstract:Accurate real-time pose estimation of spacecraft or object in space is a key capability necessary for on-orbit spacecraft servicing and assembly tasks. Pose estimation of objects in space is more challenging than for objects on Earth due to space images containing widely varying illumination conditions, high contrast, and poor resolution in addition to power and mass constraints. In this paper, a convolutional neural network is leveraged to uniquely determine the translation and rotation of an object of interest relative to the camera. The main idea of using CNN model is to assist object tracker used in on space assembly tasks where only feature based method is always not sufficient. The simulation framework designed for assembly task is used to generate dataset for training the modified CNN models and, then results of different models are compared with measure of how accurately models are predicting the pose. Unlike many current approaches for spacecraft or object in space pose estimation, the model does not rely on hand-crafted object-specific features which makes this model more robust and easier to apply to other types of spacecraft. It is shown that the model performs comparable to the current feature-selection methods and can therefore be used in conjunction with them to provide more reliable estimates.
Accurate Protein Structure Prediction by Embeddings and Deep Learning Representations
Nov 09, 2019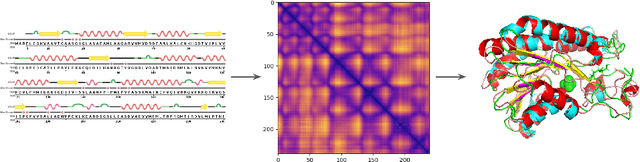

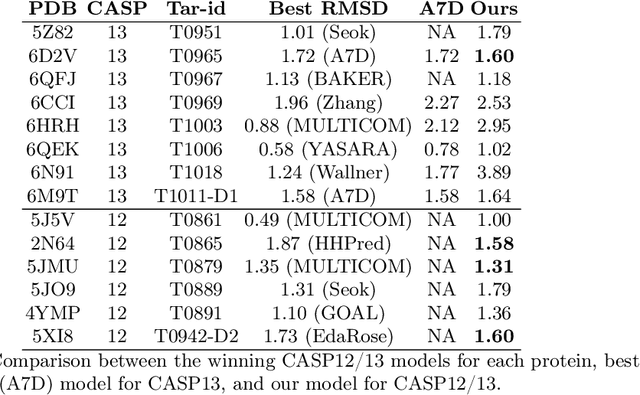
Abstract:Proteins are the major building blocks of life, and actuators of almost all chemical and biophysical events in living organisms. Their native structures in turn enable their biological functions which have a fundamental role in drug design. This motivates predicting the structure of a protein from its sequence of amino acids, a fundamental problem in computational biology. In this work, we demonstrate state-of-the-art protein structure prediction (PSP) results using embeddings and deep learning models for prediction of backbone atom distance matrices and torsion angles. We recover 3D coordinates of backbone atoms and reconstruct full atom protein by optimization. We create a new gold standard dataset of proteins which is comprehensive and easy to use. Our dataset consists of amino acid sequences, Q8 secondary structures, position specific scoring matrices, multiple sequence alignment co-evolutionary features, backbone atom distance matrices, torsion angles, and 3D coordinates. We evaluate the quality of our structure prediction by RMSD on the latest Critical Assessment of Techniques for Protein Structure Prediction (CASP) test data and demonstrate competitive results with the winning teams and AlphaFold in CASP13 and supersede the results of the winning teams in CASP12. We make our data, models, and code publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge