Jinhao Lei
Combining Compressions for Multiplicative Size Scaling on Natural Language Tasks
Aug 20, 2022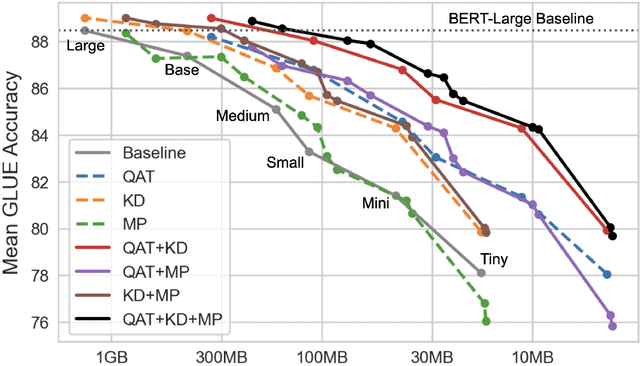
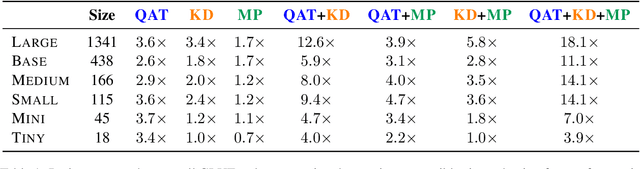
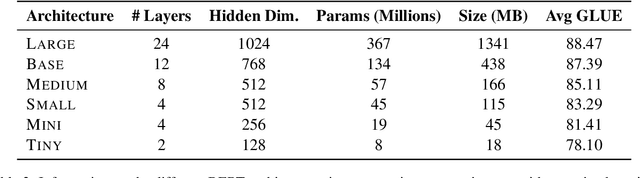
Abstract:Quantization, knowledge distillation, and magnitude pruning are among the most popular methods for neural network compression in NLP. Independently, these methods reduce model size and can accelerate inference, but their relative benefit and combinatorial interactions have not been rigorously studied. For each of the eight possible subsets of these techniques, we compare accuracy vs. model size tradeoffs across six BERT architecture sizes and eight GLUE tasks. We find that quantization and distillation consistently provide greater benefit than pruning. Surprisingly, except for the pair of pruning and quantization, using multiple methods together rarely yields diminishing returns. Instead, we observe complementary and super-multiplicative reductions to model size. Our work quantitatively demonstrates that combining compression methods can synergistically reduce model size, and that practitioners should prioritize (1) quantization, (2) knowledge distillation, and (3) pruning to maximize accuracy vs. model size tradeoffs.
Accurate Protein Structure Prediction by Embeddings and Deep Learning Representations
Nov 09, 2019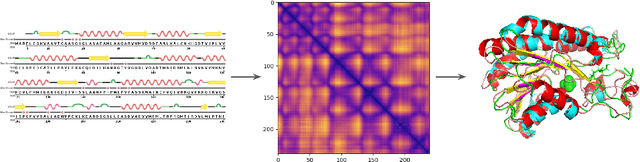
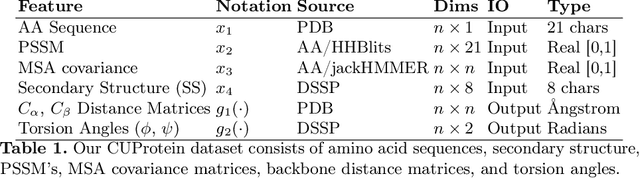

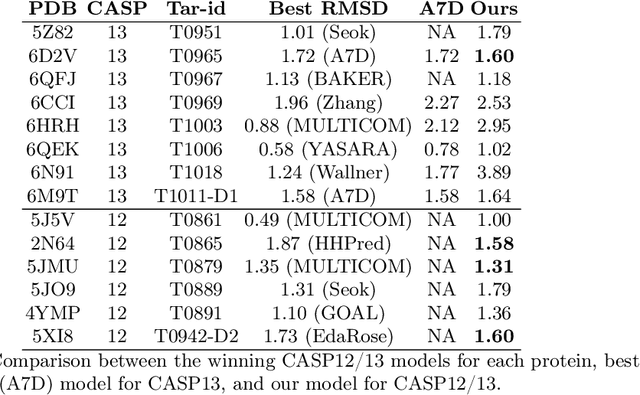
Abstract:Proteins are the major building blocks of life, and actuators of almost all chemical and biophysical events in living organisms. Their native structures in turn enable their biological functions which have a fundamental role in drug design. This motivates predicting the structure of a protein from its sequence of amino acids, a fundamental problem in computational biology. In this work, we demonstrate state-of-the-art protein structure prediction (PSP) results using embeddings and deep learning models for prediction of backbone atom distance matrices and torsion angles. We recover 3D coordinates of backbone atoms and reconstruct full atom protein by optimization. We create a new gold standard dataset of proteins which is comprehensive and easy to use. Our dataset consists of amino acid sequences, Q8 secondary structures, position specific scoring matrices, multiple sequence alignment co-evolutionary features, backbone atom distance matrices, torsion angles, and 3D coordinates. We evaluate the quality of our structure prediction by RMSD on the latest Critical Assessment of Techniques for Protein Structure Prediction (CASP) test data and demonstrate competitive results with the winning teams and AlphaFold in CASP13 and supersede the results of the winning teams in CASP12. We make our data, models, and code publicly available.
Linguistically Regularized LSTMs for Sentiment Classification
Apr 25, 2017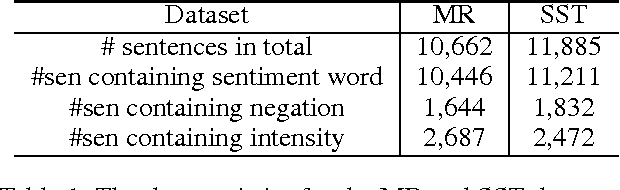
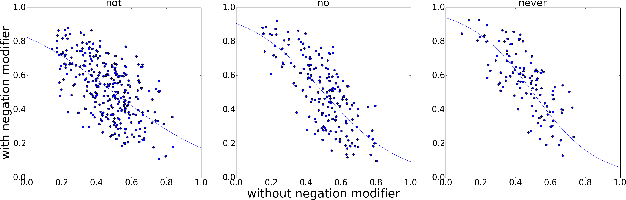
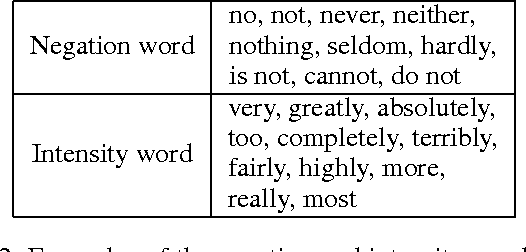
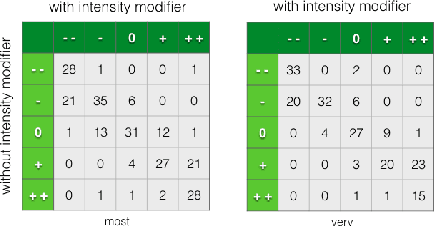
Abstract:Sentiment understanding has been a long-term goal of AI in the past decades. This paper deals with sentence-level sentiment classification. Though a variety of neural network models have been proposed very recently, however, previous models either depend on expensive phrase-level annotation, whose performance drops substantially when trained with only sentence-level annotation; or do not fully employ linguistic resources (e.g., sentiment lexicons, negation words, intensity words), thus not being able to produce linguistically coherent representations. In this paper, we propose simple models trained with sentence-level annotation, but also attempt to generating linguistically coherent representations by employing regularizers that model the linguistic role of sentiment lexicons, negation words, and intensity words. Results show that our models are effective to capture the sentiment shifting effect of sentiment, negation, and intensity words, while still obtain competitive results without sacrificing the models' simplicity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge