Dan Chen
MiLoRA: Efficient Mixture of Low-Rank Adaptation for Large Language Models Fine-tuning
Oct 23, 2024



Abstract:Low-rank adaptation (LoRA) and its mixture-of-experts (MOE) variants are highly effective parameter-efficient fine-tuning (PEFT) methods. However, they introduce significant latency in multi-tenant settings due to the LoRA modules and MOE routers added to multiple linear modules in the Transformer layer. To address this issue, we propose Mixture of Low-Rank Adaptation (MiLoRA), a novel and efficient LoRA variant. MiLoRA differs from previous MOE-style LoRA methods by considering each LoRA module as an expert and employing a prompt-aware routing mechanism. This mechanism calculates expert routing results once before generating the first new token and reuses these results for subsequent tokens, reducing latency. Extensive experiments and analysis on commonsense reasoning tasks, math reasoning tasks, and widely used LLM evaluation benchmarks demonstrate that MiLoRA consistently outperforms strong PEFT baselines with comparable tunable parameter budgets. Additionally, MiLoRA significantly reduces latency in multi-tenant settings compared to previous LoRA-based methods.
$R^3$-NL2GQL: A Hybrid Models Approach for for Accuracy Enhancing and Hallucinations Mitigation
Nov 03, 2023Abstract:While current NL2SQL tasks constructed using Foundation Models have achieved commendable results, their direct application to Natural Language to Graph Query Language (NL2GQL) tasks poses challenges due to the significant differences between GQL and SQL expressions, as well as the numerous types of GQL. Our extensive experiments reveal that in NL2GQL tasks, larger Foundation Models demonstrate superior cross-schema generalization abilities, while smaller Foundation Models struggle to improve their GQL generation capabilities through fine-tuning. However, after fine-tuning, smaller models exhibit better intent comprehension and higher grammatical accuracy. Diverging from rule-based and slot-filling techniques, we introduce R3-NL2GQL, which employs both smaller and larger Foundation Models as reranker, rewriter and refiner. The approach harnesses the comprehension ability of smaller models for information reranker and rewriter, and the exceptional generalization and generation capabilities of larger models to transform input natural language queries and code structure schema into any form of GQLs. Recognizing the lack of established datasets in this nascent domain, we have created a bilingual dataset derived from graph database documentation and some open-source Knowledge Graphs (KGs). We tested our approach on this dataset and the experimental results showed that delivers promising performance and robustness.Our code and dataset is available at https://github.com/zhiqix/NL2GQL
A Knowledge-based Learning Framework for Self-supervised Pre-training Towards Enhanced Recognition of Medical Images
Nov 27, 2022



Abstract:Self-supervised pre-training has become the priory choice to establish reliable models for automated recognition of massive medical images, which are routinely annotation-free, without semantics, and without guarantee of quality. Note that this paradigm is still at its infancy and limited by closely related open issues: 1) how to learn robust representations in an unsupervised manner from unlabelled medical images of low diversity in samples? and 2) how to obtain the most significant representations demanded by a high-quality segmentation? Aiming at these issues, this study proposes a knowledge-based learning framework towards enhanced recognition of medical images, which works in three phases by synergizing contrastive learning and generative learning models: 1) Sample Space Diversification: Reconstructive proxy tasks have been enabled to embed a priori knowledge with context highlighted to diversify the expanded sample space; 2) Enhanced Representation Learning: Informative noise-contrastive estimation loss regularizes the encoder to enhance representation learning of annotation-free images; 3) Correlated Optimization: Optimization operations in pre-training the encoder and the decoder have been correlated via image restoration from proxy tasks, targeting the need for semantic segmentation. Extensive experiments have been performed on various public medical image datasets (e.g., CheXpert and DRIVE) against the state-of-the-art counterparts (e.g., SimCLR and MoCo), and results demonstrate that: The proposed framework statistically excels in self-supervised benchmarks, achieving 2.08, 1.23, 1.12, 0.76 and 1.38 percentage points improvements over SimCLR in AUC/Dice. The proposed framework achieves label-efficient semi-supervised learning, e.g., reducing the annotation cost by up to 99% in pathological classification.
From WSI-level to Patch-level: Structure Prior Guided Binuclear Cell Fine-grained Detection
Aug 26, 2022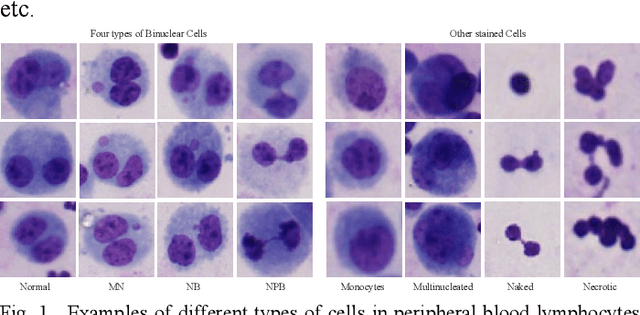
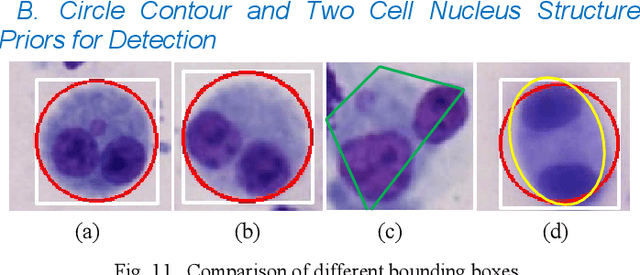
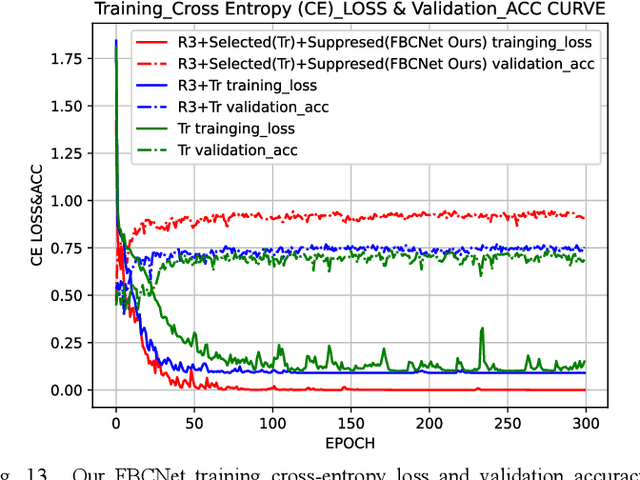
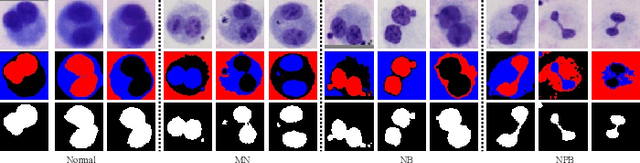
Abstract:Accurately and quickly binuclear cell (BC) detection plays a significant role in predicting the risk of leukemia and other malignant tumors. However, manual microscopy counting is time-consuming and lacks objectivity. Moreover, with the limitation of staining quality and diversity of morphology features in BC microscopy whole slide images (WSIs), traditional image processing approaches are helpless. To overcome this challenge, we propose a two-stage detection method inspired by the structure prior of BC based on deep learning, which cascades to implement BCs coarse detection at the WSI-level and fine-grained classification in patch-level. The coarse detection network is a multi-task detection framework based on circular bounding boxes for cells detection, and central key points for nucleus detection. The circle representation reduces the degrees of freedom, mitigates the effect of surrounding impurities compared to usual rectangular boxes and can be rotation invariant in WSI. Detecting key points in the nucleus can assist network perception and be used for unsupervised color layer segmentation in later fine-grained classification. The fine classification network consists of a background region suppression module based on color layer mask supervision and a key region selection module based on a transformer due to its global modeling capability. Additionally, an unsupervised and unpaired cytoplasm generator network is firstly proposed to expand the long-tailed distribution dataset. Finally, experiments are performed on BC multicenter datasets. The proposed BC fine detection method outperforms other benchmarks in almost all the evaluation criteria, providing clarification and support for tasks such as cancer screenings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge