Daiwei Lu
EndoStreamDepth: Temporally Consistent Monocular Depth Estimation for Endoscopic Video Streams
Dec 20, 2025



Abstract:This work presents EndoStreamDepth, a monocular depth estimation framework for endoscopic video streams. It provides accurate depth maps with sharp anatomical boundaries for each frame, temporally consistent predictions across frames, and real-time throughput. Unlike prior work that uses batched inputs, EndoStreamDepth processes individual frames with a temporal module to propagate inter-frame information. The framework contains three main components: (1) a single-frame depth network with endoscopy-specific transformation to produce accurate depth maps, (2) multi-level Mamba temporal modules that leverage inter-frame information to improve accuracy and stabilize predictions, and (3) a hierarchical design with comprehensive multi-scale supervision, where complementary loss terms jointly improve local boundary sharpness and global geometric consistency. We conduct comprehensive evaluations on two publicly available colonoscopy depth estimation datasets. Compared to state-of-the-art monocular depth estimation methods, EndoStreamDepth substantially improves performance, and it produces depth maps with sharp, anatomically aligned boundaries, which are essential to support downstream tasks such as automation for robotic surgery. The code is publicly available at https://github.com/MedICL-VU/EndoStreamDepth
Endo-SemiS: Towards Robust Semi-Supervised Image Segmentation for Endoscopic Video
Dec 18, 2025



Abstract:In this paper, we present Endo-SemiS, a semi-supervised segmentation framework for providing reliable segmentation of endoscopic video frames with limited annotation. EndoSemiS uses 4 strategies to improve performance by effectively utilizing all available data, particularly unlabeled data: (1) Cross-supervision between two individual networks that supervise each other; (2) Uncertainty-guided pseudo-labels from unlabeled data, which are generated by selecting high-confidence regions to improve their quality; (3) Joint pseudolabel supervision, which aggregates reliable pixels from the pseudo-labels of both networks to provide accurate supervision for unlabeled data; and (4) Mutual learning, where both networks learn from each other at the feature and image levels, reducing variance and guiding them toward a consistent solution. Additionally, a separate corrective network that utilizes spatiotemporal information from endoscopy video to improve segmentation performance. Endo-SemiS is evaluated on two clinical applications: kidney stone laser lithotomy from ureteroscopy and polyp screening from colonoscopy. Compared to state-of-the-art segmentation methods, Endo-SemiS substantially achieves superior results on both datasets with limited labeled data. The code is publicly available at https://github.com/MedICL-VU/Endo-SemiS
SynStitch: a Self-Supervised Learning Network for Ultrasound Image Stitching Using Synthetic Training Pairs and Indirect Supervision
Nov 11, 2024
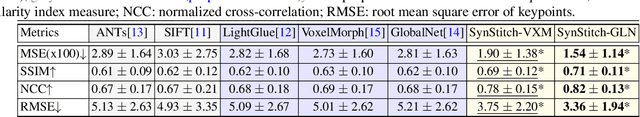
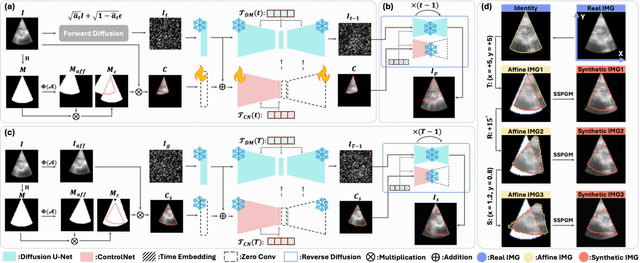

Abstract:Ultrasound (US) image stitching can expand the field-of-view (FOV) by combining multiple US images from varied probe positions. However, registering US images with only partially overlapping anatomical contents is a challenging task. In this work, we introduce SynStitch, a self-supervised framework designed for 2DUS stitching. SynStitch consists of a synthetic stitching pair generation module (SSPGM) and an image stitching module (ISM). SSPGM utilizes a patch-conditioned ControlNet to generate realistic 2DUS stitching pairs with known affine matrix from a single input image. ISM then utilizes this synthetic paired data to learn 2DUS stitching in a supervised manner. Our framework was evaluated against multiple leading methods on a kidney ultrasound dataset, demonstrating superior 2DUS stitching performance through both qualitative and quantitative analyses. The code will be made public upon acceptance of the paper.
AdaptDiff: Cross-Modality Domain Adaptation via Weak Conditional Semantic Diffusion for Retinal Vessel Segmentation
Oct 06, 2024Abstract:Deep learning has shown remarkable performance in medical image segmentation. However, despite its promise, deep learning has many challenges in practice due to its inability to effectively transition to unseen domains, caused by the inherent data distribution shift and the lack of manual annotations to guide domain adaptation. To tackle this problem, we present an unsupervised domain adaptation (UDA) method named AdaptDiff that enables a retinal vessel segmentation network trained on fundus photography (FP) to produce satisfactory results on unseen modalities (e.g., OCT-A) without any manual labels. For all our target domains, we first adopt a segmentation model trained on the source domain to create pseudo-labels. With these pseudo-labels, we train a conditional semantic diffusion probabilistic model to represent the target domain distribution. Experimentally, we show that even with low quality pseudo-labels, the diffusion model can still capture the conditional semantic information. Subsequently, we sample on the target domain with binary vessel masks from the source domain to get paired data, i.e., target domain synthetic images conditioned on the binary vessel map. Finally, we fine-tune the pre-trained segmentation network using the synthetic paired data to mitigate the domain gap. We assess the effectiveness of AdaptDiff on seven publicly available datasets across three distinct modalities. Our results demonstrate a significant improvement in segmentation performance across all unseen datasets. Our code is publicly available at https://github.com/DeweiHu/AdaptDiff.
False Negative/Positive Control for SAM on Noisy Medical Images
Aug 20, 2023Abstract:The Segment Anything Model (SAM) is a recently developed all-range foundation model for image segmentation. It can use sparse manual prompts such as bounding boxes to generate pixel-level segmentation in natural images but struggles in medical images such as low-contrast, noisy ultrasound images. We propose a refined test-phase prompt augmentation technique designed to improve SAM's performance in medical image segmentation. The method couples multi-box prompt augmentation and an aleatoric uncertainty-based false-negative (FN) and false-positive (FP) correction (FNPC) strategy. We evaluate the method on two ultrasound datasets and show improvement in SAM's performance and robustness to inaccurate prompts, without the necessity for further training or tuning. Moreover, we present the Single-Slice-to-Volume (SS2V) method, enabling 3D pixel-level segmentation using only the bounding box annotation from a single 2D slice. Our results allow efficient use of SAM in even noisy, low-contrast medical images. The source code will be released soon.
SSL^2: Self-Supervised Learning meets Semi-Supervised Learning: Multiple Sclerosis Segmentation in 7T-MRI from large-scale 3T-MRI
Mar 09, 2023Abstract:Automated segmentation of multiple sclerosis (MS) lesions from MRI scans is important to quantify disease progression. In recent years, convolutional neural networks (CNNs) have shown top performance for this task when a large amount of labeled data is available. However, the accuracy of CNNs suffers when dealing with few and/or sparsely labeled datasets. A potential solution is to leverage the information available in large public datasets in conjunction with a target dataset which only has limited labeled data. In this paper, we propose a training framework, SSL2 (self-supervised-semi-supervised), for multi-modality MS lesion segmentation with limited supervision. We adopt self-supervised learning to leverage the knowledge from large public 3T datasets to tackle the limitations of a small 7T target dataset. To leverage the information from unlabeled 7T data, we also evaluate state-of-the-art semi-supervised methods for other limited annotation settings, such as small labeled training size and sparse annotations. We use the shifted-window (Swin) transformer1 as our backbone network. The effectiveness of self-supervised and semi-supervised training strategies is evaluated in our in-house 7T MRI dataset. The results indicate that each strategy improves lesion segmentation for both limited training data size and for sparse labeling scenarios. The combined overall framework further improves the performance substantially compared to either of its components alone. Our proposed framework thus provides a promising solution for future data/label-hungry 7T MS studies.
Segmentation of kidney stones in endoscopic video feeds
Apr 29, 2022



Abstract:Image segmentation has been increasingly applied in medical settings as recent developments have skyrocketed the potential applications of deep learning. Urology, specifically, is one field of medicine that is primed for the adoption of a real-time image segmentation system with the long-term aim of automating endoscopic stone treatment. In this project, we explored supervised deep learning models to annotate kidney stones in surgical endoscopic video feeds. In this paper, we describe how we built a dataset from the raw videos and how we developed a pipeline to automate as much of the process as possible. For the segmentation task, we adapted and analyzed three baseline deep learning models -- U-Net, U-Net++, and DenseNet -- to predict annotations on the frames of the endoscopic videos with the highest accuracy above 90\%. To show clinical potential for real-time use, we also confirmed that our best trained model can accurately annotate new videos at 30 frames per second. Our results demonstrate that the proposed method justifies continued development and study of image segmentation to annotate ureteroscopic video feeds.
* Published in SPIE Medical Imaging: Image Processing 2022 (9 pages, 5 figures, 1 table)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge