Clement Vignac
Generative Modelling of Structurally Constrained Graphs
Jun 25, 2024


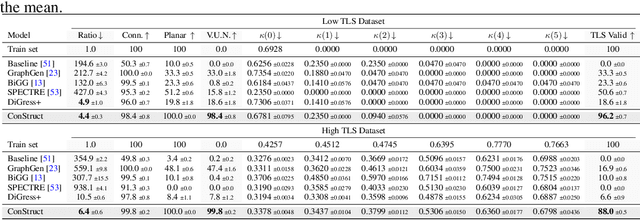
Abstract:Graph diffusion models have emerged as state-of-the-art techniques in graph generation, yet integrating domain knowledge into these models remains challenging. Domain knowledge is particularly important in real-world scenarios, where invalid generated graphs hinder deployment in practical applications. Unconstrained and conditioned graph generative models fail to guarantee such domain-specific structural properties. We present ConStruct, a novel framework that allows for hard-constraining graph diffusion models to incorporate specific properties, such as planarity or acyclicity. Our approach ensures that the sampled graphs remain within the domain of graphs that verify the specified property throughout the entire trajectory in both the forward and reverse processes. This is achieved by introducing a specific edge-absorbing noise model and a new projector operator. ConStruct demonstrates versatility across several structural and edge-deletion invariant constraints and achieves state-of-the-art performance for both synthetic benchmarks and attributed real-world datasets. For example, by leveraging planarity in digital pathology graph datasets, the proposed method outperforms existing baselines and enhances generated data validity by up to 71.1 percentage points.
Sparse Training of Discrete Diffusion Models for Graph Generation
Nov 03, 2023



Abstract:Generative models for graphs often encounter scalability challenges due to the inherent need to predict interactions for every node pair. Despite the sparsity often exhibited by real-world graphs, the unpredictable sparsity patterns of their adjacency matrices, stemming from their unordered nature, leads to quadratic computational complexity. In this work, we introduce SparseDiff, a denoising diffusion model for graph generation that is able to exploit sparsity during its training phase. At the core of SparseDiff is a message-passing neural network tailored to predict only a subset of edges during each forward pass. When combined with a sparsity-preserving noise model, this model can efficiently work with edge lists representations of graphs, paving the way for scalability to much larger structures. During the sampling phase, SparseDiff iteratively populates the adjacency matrix from its prior state, ensuring prediction of the full graph while controlling memory utilization. Experimental results show that SparseDiff simultaneously matches state-of-the-art in generation performance on both small and large graphs, highlighting the versatility of our method.
MiDi: Mixed Graph and 3D Denoising Diffusion for Molecule Generation
Feb 17, 2023



Abstract:This work introduces MiDi, a diffusion model for jointly generating molecular graphs and corresponding 3D conformers. In contrast to existing models, which derive molecular bonds from the conformation using predefined rules, MiDi streamlines the molecule generation process with an end-to-end differentiable model. Experimental results demonstrate the benefits of this approach: on the complex GEOM-DRUGS dataset, our model generates significantly better molecular graphs than 3D-based models and even surpasses specialized algorithms that directly optimize the bond orders for validity. Our code is available at github.com/cvignac/MiDi.
DiGress: Discrete Denoising diffusion for graph generation
Sep 29, 2022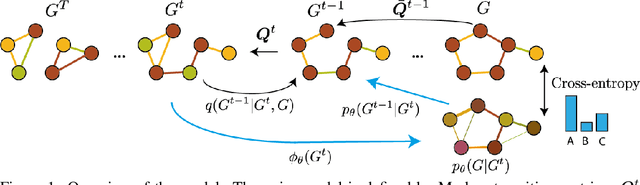
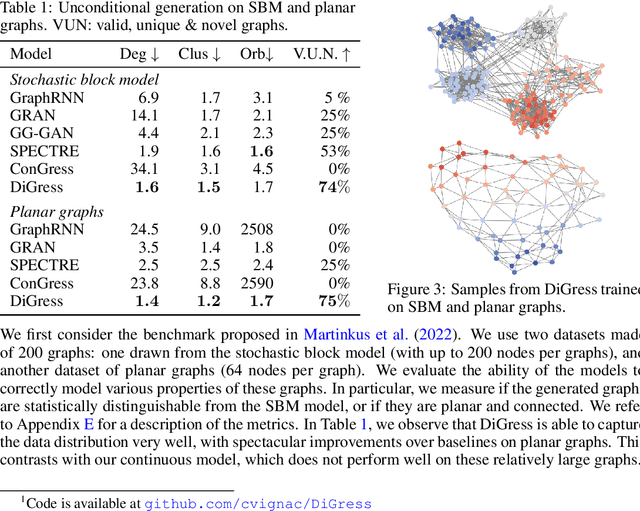
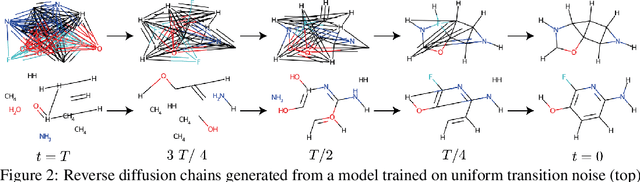
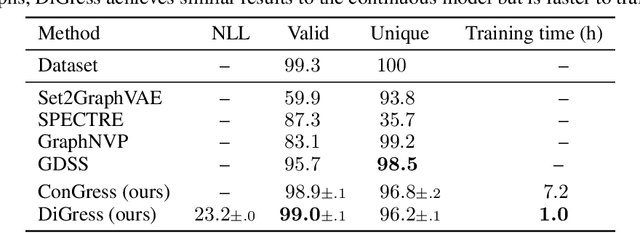
Abstract:This work introduces DiGress, a discrete denoising diffusion model for generating graphs with categorical node and edge attributes. Our model defines a diffusion process that progressively edits a graph with noise (adding or removing edges, changing the categories), and a graph transformer network that learns to revert this process. With these two ingredients in place, we reduce distribution learning over graphs to a simple sequence of classification tasks. We further improve sample quality by proposing a new Markovian noise model that preserves the marginal distribution of node and edge types during diffusion, and by adding auxiliary graph-theoretic features derived from the noisy graph at each diffusion step. Finally, we propose a guidance procedure for conditioning the generation on graph-level features. Overall, DiGress achieves state-of-the-art performance on both molecular and non-molecular datasets, with up to 3x validity improvement on a dataset of planar graphs. In particular, it is the first model that scales to the large GuacaMol dataset containing 1.3M drug-like molecules without using a molecule-specific representation such as SMILES or fragments.
Top-N: Equivariant set and graph generation without exchangeability
Oct 05, 2021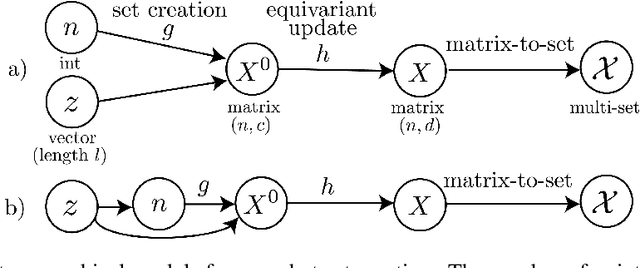

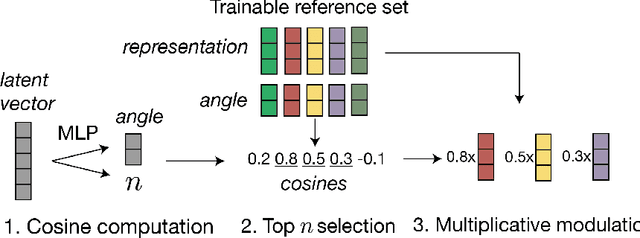

Abstract:We consider one-shot probabilistic decoders that map a vector-shaped prior to a distribution over sets or graphs. These functions can be integrated into variational autoencoders (VAE), generative adversarial networks (GAN) or normalizing flows, and have important applications in drug discovery. Set and graph generation is most commonly performed by generating points (and sometimes edge weights) i.i.d. from a normal distribution, and processing them along with the prior vector using Transformer layers or graph neural networks. This architecture is designed to generate exchangeable distributions (all permutations of a set are equally likely) but it is hard to train due to the stochasticity of i.i.d. generation. We propose a new definition of equivariance and show that exchangeability is in fact unnecessary in VAEs and GANs. We then introduce Top-n, a deterministic, non-exchangeable set creation mechanism which learns to select the most relevant points from a trainable reference set. Top-n can replace i.i.d. generation in any VAE or GAN -- it is easier to train and better captures complex dependencies in the data. Top-n outperforms i.i.d generation by 15% at SetMNIST reconstruction, generates sets that are 64% closer to the true distribution on a synthetic molecule-like dataset, and is able to generate more diverse molecules when trained on the classical QM9 dataset. With improved foundations in one-shot generation, our algorithm contributes to the design of more effective molecule generation methods.
Building powerful and equivariant graph neural networks with structural message-passing
Jul 11, 2020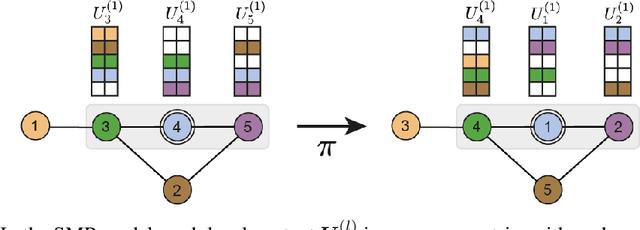
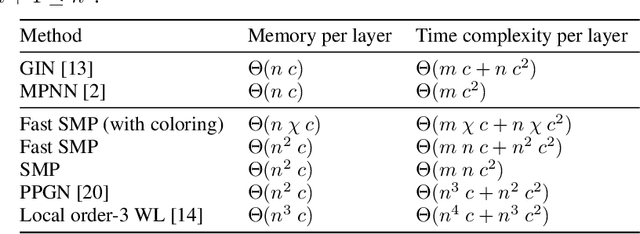

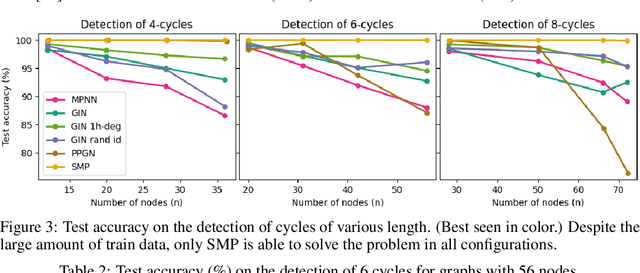
Abstract:Message-passing has proved to be an effective way to design graph neural networks, as it is able to leverage both permutation equivariance and an inductive bias towards learning local structures to achieve good generalization. However, current message-passing architectures have a limited representation power and fail to learn basic topological properties of graphs. We address this problem and propose a new message-passing framework that is powerful while preserving permutation equivariance. Specifically, we propagate unique node identifiers in the form of a one-hot encoding in order to learn a local context matrix around each node. This enables to learn rich local information about both features and topology, which can be pooled to obtain node representations. Experimentally, we find our model to be superior at predicting various graph topological properties, opening the way to novel powerful architectures that are both equivariant and computationally efficient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge