Chowdhury Rafeed Rahman
How much can ChatGPT really help Computational Biologists in Programming?
Sep 17, 2023Abstract:ChatGPT, a recently developed product by openAI, is successfully leaving its mark as a multi-purpose natural language based chatbot. In this paper, we are more interested in analyzing its potential in the field of computational biology. A major share of work done by computational biologists these days involve coding up Bioinformatics algorithms, analyzing data, creating pipelining scripts and even machine learning modeling & feature extraction. This paper focuses on the potential influence (both positive and negative) of ChatGPT in the mentioned aspects with illustrative examples from different perspectives. Compared to other fields of Computer Science, Computational Biology has: (1) less coding resources, (2) more sensitivity and bias issues (deals with medical data) and (3) more necessity of coding assistance (people from diverse background come to this field). Keeping such issues in mind, we cover use cases such as code writing, reviewing, debugging, converting, refactoring and pipelining using ChatGPT from the perspective of computational biologists in this paper.
BSpell: A CNN-blended BERT Based Bengali Spell Checker
Aug 20, 2022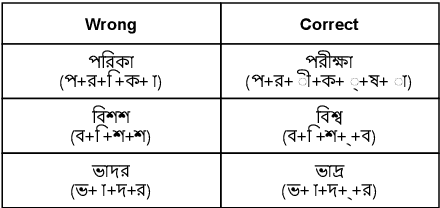
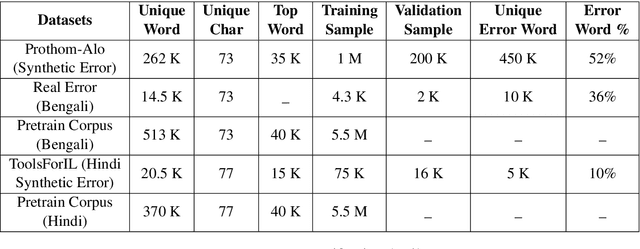

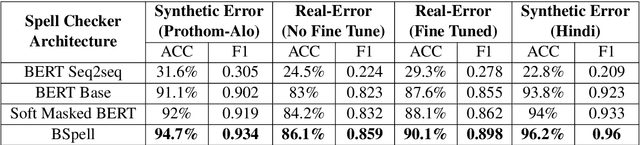
Abstract:Bengali typing is mostly performed using English keyboard and can be highly erroneous due to the presence of compound and similarly pronounced letters. Spelling correction of a misspelled word requires understanding of word typing pattern as well as the context of the word usage. We propose a specialized BERT model, BSpell targeted towards word for word correction in sentence level. BSpell contains an end-to-end trainable CNN sub-model named SemanticNet along with specialized auxiliary loss. This allows BSpell to specialize in highly inflected Bengali vocabulary in the presence of spelling errors. We further propose hybrid pretraining scheme for BSpell combining word level and character level masking. Utilizing this pretraining scheme, BSpell achieves 91.5% accuracy on real life Bengali spelling correction validation set. Detailed comparison on two Bengali and one Hindi spelling correction dataset shows the superiority of proposed BSpell over existing spell checkers.
Judge a Sentence by Its Content to Generate Grammatical Errors
Aug 20, 2022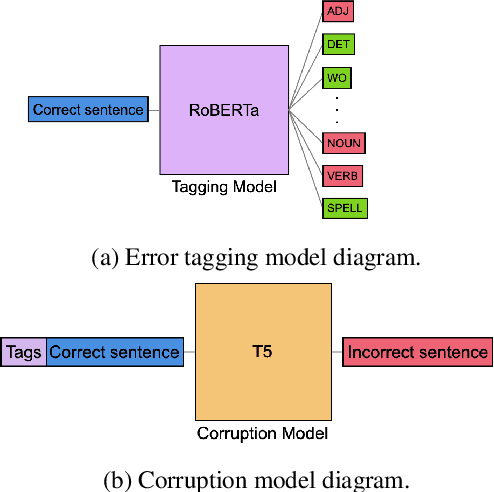
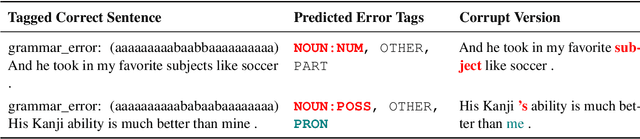
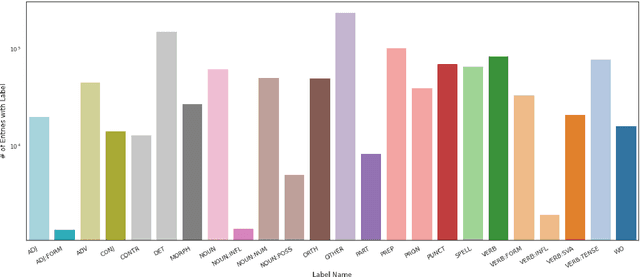
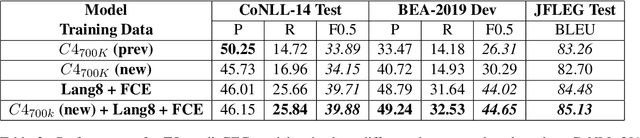
Abstract:Data sparsity is a well-known problem for grammatical error correction (GEC). Generating synthetic training data is one widely proposed solution to this problem, and has allowed models to achieve state-of-the-art (SOTA) performance in recent years. However, these methods often generate unrealistic errors, or aim to generate sentences with only one error. We propose a learning based two stage method for synthetic data generation for GEC that relaxes this constraint on sentences containing only one error. Errors are generated in accordance with sentence merit. We show that a GEC model trained on our synthetically generated corpus outperforms models trained on synthetic data from prior work.
Paradigm Shift in Language Modeling: Revisiting CNN for Modeling Sanskrit Originated Bengali and Hindi Language
Nov 05, 2021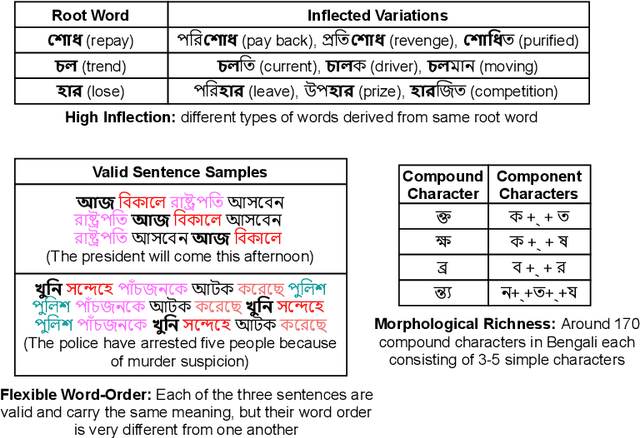
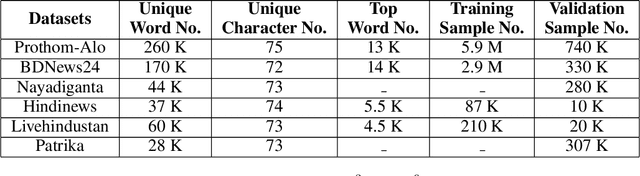
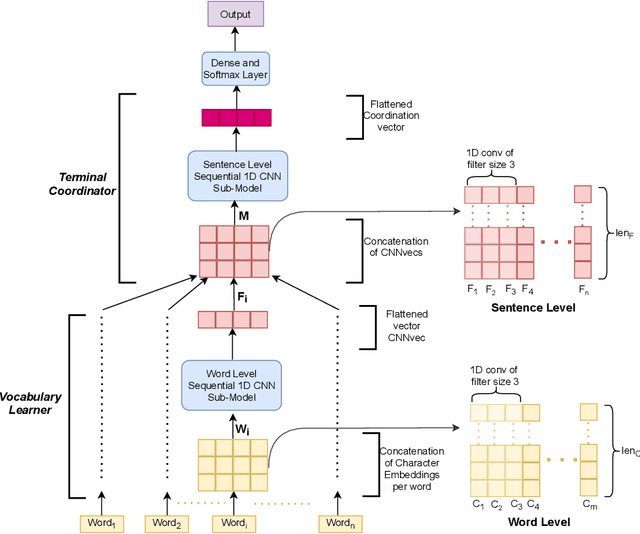
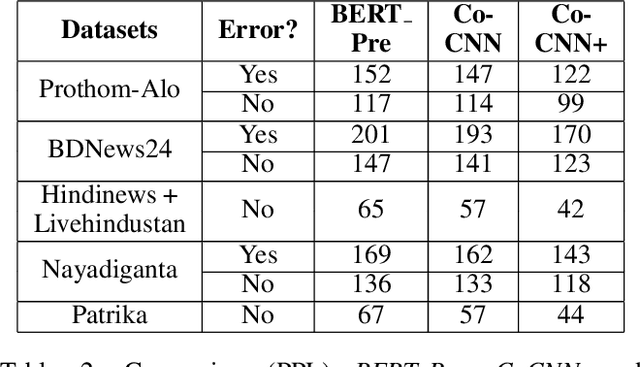
Abstract:Though there has been a large body of recent works in language modeling (LM) for high resource languages such as English and Chinese, the area is still unexplored for low resource languages like Bengali and Hindi. We propose an end to end trainable memory efficient CNN architecture named CoCNN to handle specific characteristics such as high inflection, morphological richness, flexible word order and phonetical spelling errors of Bengali and Hindi. In particular, we introduce two learnable convolutional sub-models at word and at sentence level that are end to end trainable. We show that state-of-the-art (SOTA) Transformer models including pretrained BERT do not necessarily yield the best performance for Bengali and Hindi. CoCNN outperforms pretrained BERT with 16X less parameters, and it achieves much better performance than SOTA LSTM models on multiple real-world datasets. This is the first study on the effectiveness of different architectures drawn from three deep learning paradigms - Convolution, Recurrent, and Transformer neural nets for modeling two widely used languages, Bengali and Hindi.
i6mA-CNN: a convolution based computational approach towards identification of DNA N6-methyladenine sites in rice genome
Aug 11, 2020



Abstract:DNA N6-methylation (6mA) in Adenine nucleotide is a post replication modification and is responsible for many biological functions. Experimental methods for genome wide 6mA site detection is an expensive and manual labour intensive process. Automated and accurate computational methods can help to identify 6mA sites in long genomes saving significant time and money. Our study develops a convolutional neural network based tool i6mA-CNN capable of identifying 6mA sites in the rice genome. Our model coordinates among multiple types of features such as PseAAC inspired customized feature vector, multiple one hot representations and dinucleotide physicochemical properties. It achieves area under the receiver operating characteristic curve of 0.98 with an overall accuracy of 0.94 using 5 fold cross validation on benchmark dataset. Finally, we evaluate our model on two other plant genome 6mA site identification datasets besides rice. Results suggest that our proposed tool is able to generalize its ability of 6mA site identification on plant genomes irrespective of plant species. Web tool for this research can be found at: https://cutt.ly/Co6KuWG. Supplementary data (benchmark dataset, independent test dataset, comparison purpose dataset, trained model, physicochemical property values, attention mechanism details for motif finding) are available at https://cutt.ly/PpDdeDH.
Rice grain disease identification using dual phase convolutional neural network-based system aimed at small dataset
Apr 21, 2020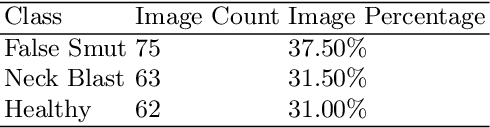

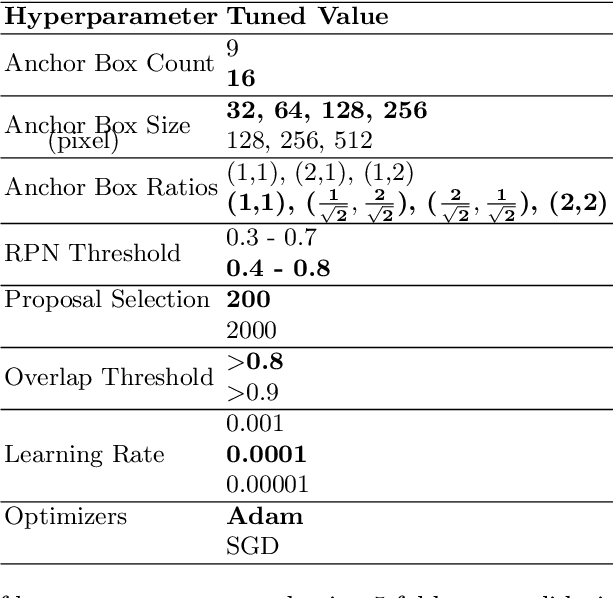
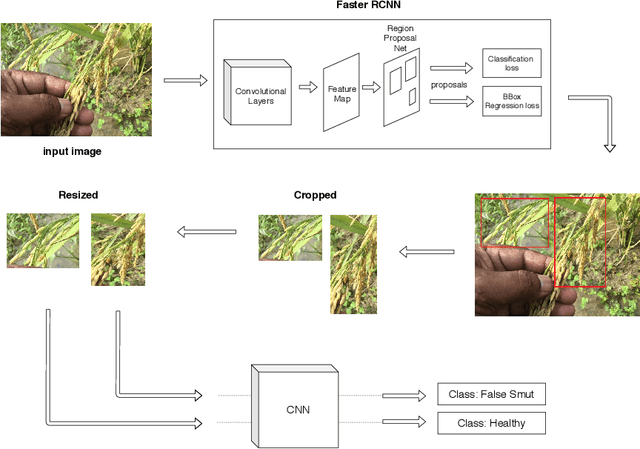
Abstract:Although Convolutional neural networks (CNNs) are widely used for plant disease detection, they require a large number of training samples when dealing with wide variety of heterogeneous background. In this work, a CNN based dual phase method has been proposed which can work effectively on small rice grain disease dataset with heterogeneity. At the first phase, Faster RCNN method is applied for cropping out the significant portion (rice grain) from the image. This initial phase results in a secondary dataset of rice grains devoid of heterogeneous background. Disease classification is performed on such derived and simplified samples using CNN architecture. Comparison of the dual phase approach with straight forward application of CNN on the small grain dataset shows the effectiveness of the proposed method which provides a 5 fold cross validation accuracy of 88.07%.
Constraints in Developing a Complete Bengali Optical Character Recognition System
Mar 22, 2020



Abstract:Technological advancement has led to digitizing hard copies of media effortlessly with optical character recognition (OCR) system. As OCR systems are being used constantly, converting printed or handwritten documents and books have become simple and time efficient. To be a fully functional structure, Bengali OCR system needs to overcome some constraints involved in pre-processing, segmentation and recognition phase. The aim of this research is to analyze the challenges prevalent in developing a Bengali OCR system through robust literature review and implementation.
Synthetic Error Dataset Generation Mimicking Bengali Writing Pattern
Mar 07, 2020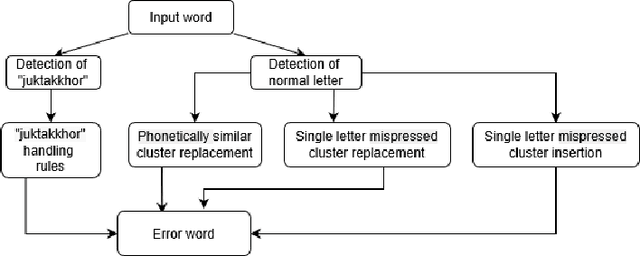
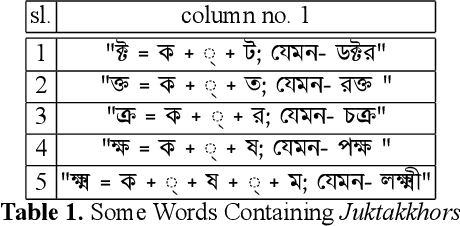
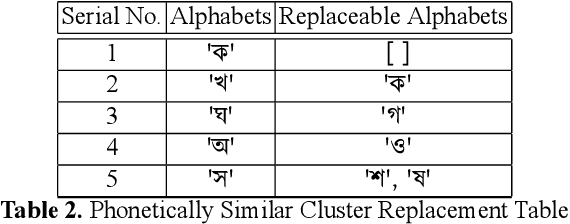
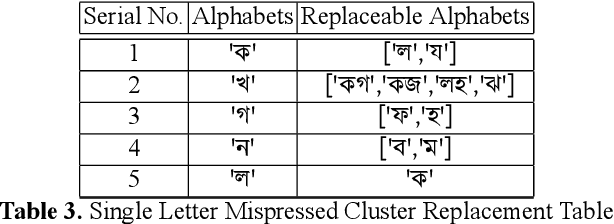
Abstract:While writing Bengali using English keyboard, users often make spelling mistakes. The accuracy of any Bengali spell checker or paragraph correction module largely depends on the kind of error dataset it is based on. Manual generation of such error dataset is a cumbersome process. In this research, We present an algorithm for automatic misspelled Bengali word generation from correct word through analyzing Bengali writing pattern using QWERTY layout English keyboard. As part of our analysis, we have formed a list of most commonly used Bengali words, phonetically similar replaceable clusters, frequently mispressed replaceable clusters, frequently mispressed insertion prone clusters and some rules for Juktakkhar (constant letter clusters) handling while generating errors.
Automatic Signboard Detection from Natural Scene Image in Context of Bangladesh Google Street View
Mar 06, 2020

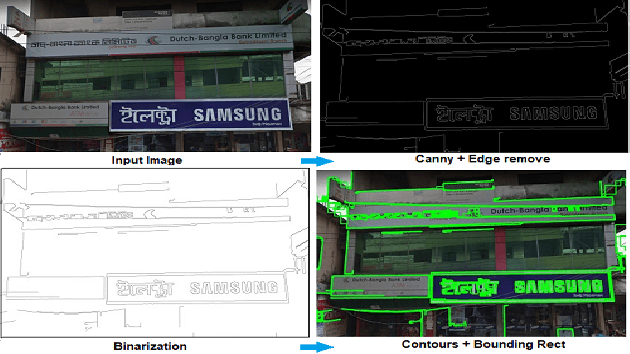

Abstract:Automatic signboard region detection is the first step of information extraction about establishments from an image, especially when there is a complex background and multiple signboard regions are present in the image. Automatic signboard detection in Bangladesh is a challenging task because of low quality street view image, presence of overlapping objects and presence of signboard like objects which are not actually signboards. In this research, we provide a novel dataset from the perspective of Bangladesh city streets with an aim of signboard detection, namely Bangladesh Street View Signboard Objects (BSVSO) image dataset. We introduce a novel approach to detect signboard accurately by applying smart image processing techniques and statistically determined hyperparameter based deep learning method, Faster R-CNN. Comparison of different variations of this segmentation based learning method have also been performed in this research.
iPromoter-BnCNN: a Novel Branched CNN Based Predictor for Identifying and Classifying Sigma Promoters
Jan 10, 2020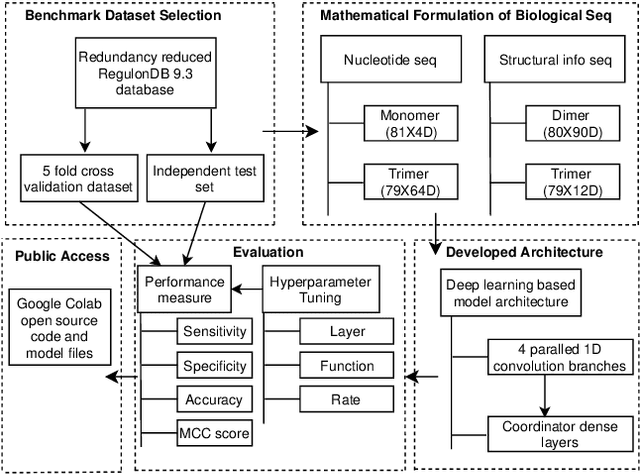
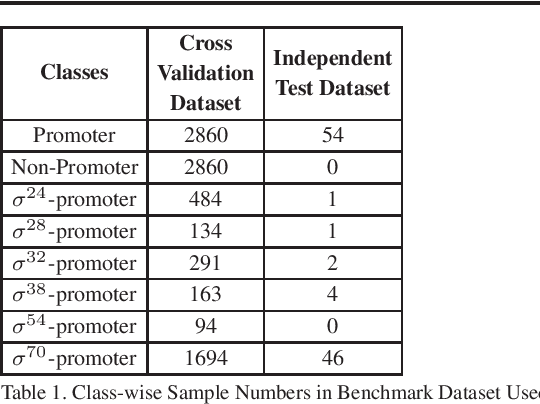
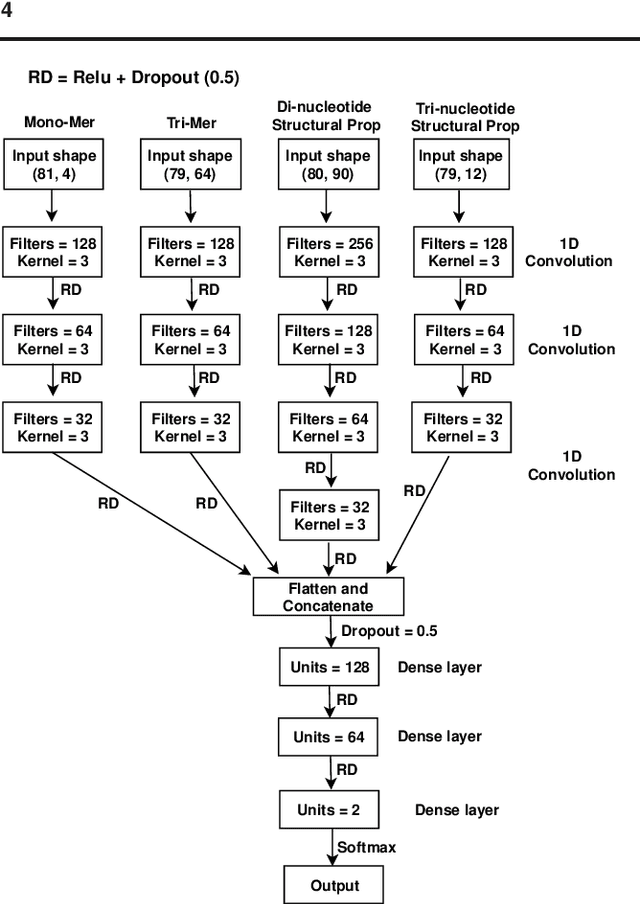
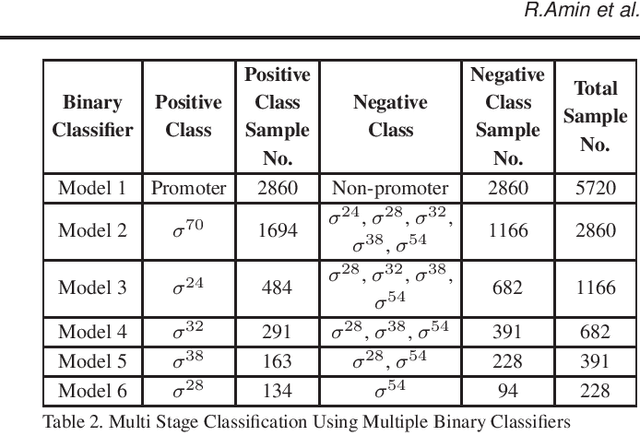
Abstract:Promoter is a short region of DNA which is responsible for initiating transcription of specific genes. Development of computational tools for automatic identification of promoters is in high demand. According to the difference of functions, promoters can be of different types. Promoters may have both intra and inter class variation and similarity in terms of consensus sequences. Accurate classification of various types of sigma promoters still remains a challenge. We present iPromoter-BnCNN for identification and accurate classification of six types of promoters - sigma24, sigma28, sigma32, sigma38, sigma54, sigma70. It is a Convolutional Neural Network (CNN) based classifier which combines local features related to monomer nucleotide sequence, trimer nucleotide sequence, dimer structural properties and trimer structural properties through the use of parallel branching. We conducted experiments on a benchmark dataset and compared with two state-of-the-art tools to show our supremacy on 5-fold cross-validation. Moreover, we tested our classifier on an independent test dataset. Our proposed tool iPromoter-BnCNN along with the source code is freely available at https://cutt.ly/te6XISV.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge