Briton Park
An Investigation into the Effects of Pre-training Data Distributions for Pathology Report Classification
May 27, 2023



Abstract:Pre-trained transformer models have demonstrated success across many natural language processing (NLP) tasks. In applying these models to the clinical domain, a prevailing assumption is that pre-training language models from scratch on large-scale biomedical data results in substantial improvements. We test this assumption with 4 pathology classification tasks on a corpus of 2907 prostate cancer pathology reports. We evaluate 5 transformer pre-trained models that are the same size but differ in pre-training corpora. Specifically, we analyze 3 categories of models: 1)General-domain: BERT and Turing Natural Language Representation (TNLR) models, which use general corpora for pre-training, 2)Mixed-domain: BioBERT which is obtained from BERT by including PubMed abstracts in pre-training and Clinical BioBERT which additionally includes MIMIC-III clinical notes and 3)Domain-specific: PubMedBERT which is pre-trained from scratch on PubMed abstracts. We find the mixed-domain and domain-specific models exhibit faster feature disambiguation during fine-tuning. However, the domain-specific model, PubMedBERT, can overfit to minority classes when presented with class imbalance, a common scenario in pathology report data. At the same time, the mixed-domain models are more resistant to overfitting. Our findings indicate that the use of general natural language and domain-specific corpora in pre-training serve complementary purposes for pathology report classification. The first enables resistance to overfitting when fine-tuning on an imbalanced dataset while the second allows for more accurate modelling of the fine-tuning domain. An expert evaluation is also conducted to reveal common outlier modes of each model. Our results could inform better fine-tuning practices in the clinical domain, to possibly leverage the benefits of mixed-domain models for imbalanced downstream datasets.
Enriched Annotations for Tumor Attribute Classification from Pathology Reports with Limited Labeled Data
Dec 15, 2020



Abstract:Precision medicine has the potential to revolutionize healthcare, but much of the data for patients is locked away in unstructured free-text, limiting research and delivery of effective personalized treatments. Generating large annotated datasets for information extraction from clinical notes is often challenging and expensive due to the high level of expertise needed for high quality annotations. To enable natural language processing for small dataset sizes, we develop a novel enriched hierarchical annotation scheme and algorithm, Supervised Line Attention (SLA), and apply this algorithm to predicting categorical tumor attributes from kidney and colon cancer pathology reports from the University of California San Francisco (UCSF). Whereas previous work only annotated document level labels, we in addition ask the annotators to enrich the traditional label by asking them to also highlight the relevant line or potentially lines for the final label, which leads to a 20% increase of annotation time required per document. With the enriched annotations, we develop a simple and interpretable machine learning algorithm that first predicts the relevant lines in the document and then predicts the tumor attribute. Our results show across the small dataset sizes of 32, 64, 128, and 186 labeled documents per cancer, SLA only requires half the number of labeled documents as state-of-the-art methods to achieve similar or better micro-f1 and macro-f1 scores for the vast majority of comparisons that we made. Accounting for the increased annotation time, this leads to a 40% reduction in total annotation time over the state of the art.
Stable discovery of interpretable subgroups via calibration in causal studies
Sep 29, 2020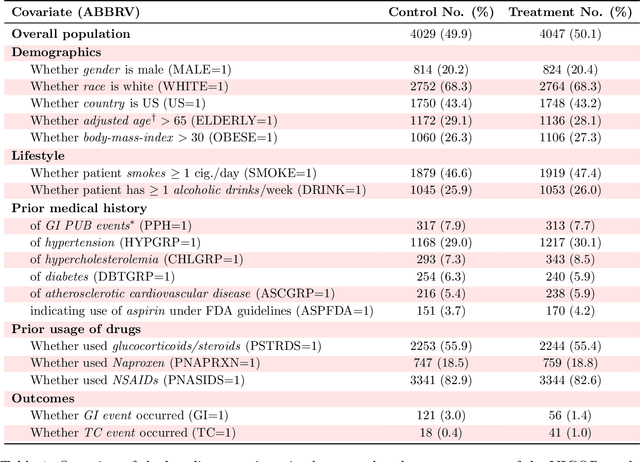
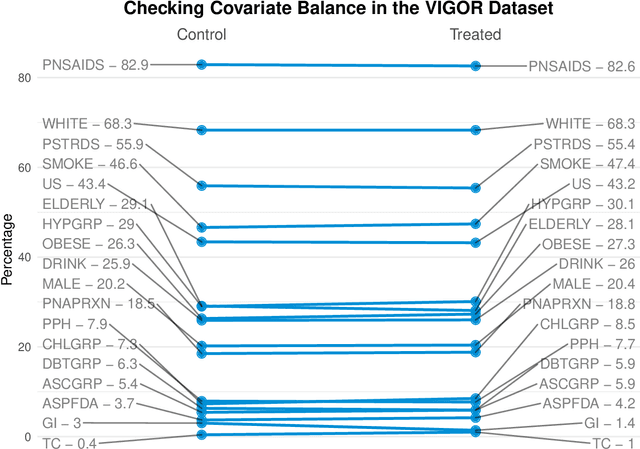
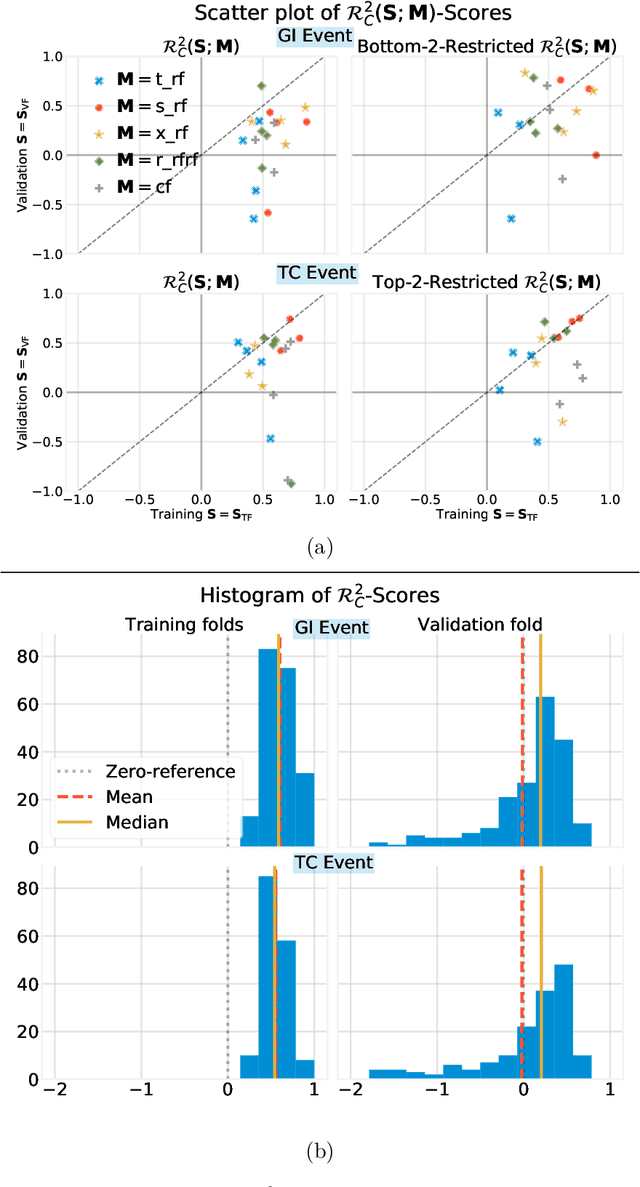
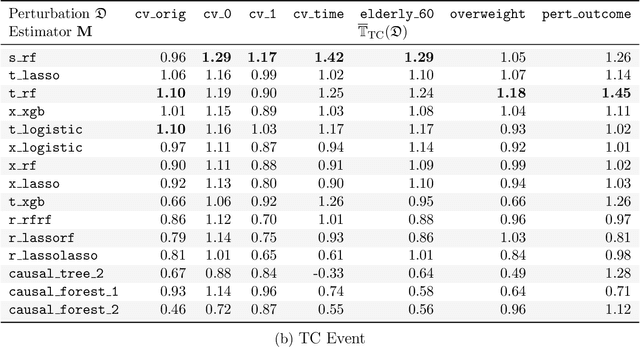
Abstract:Building on Yu and Kumbier's PCS framework and for randomized experiments, we introduce a novel methodology for Stable Discovery of Interpretable Subgroups via Calibration (StaDISC), with large heterogeneous treatment effects. StaDISC was developed during our re-analysis of the 1999-2000 VIGOR study, an 8076 patient randomized controlled trial (RCT), that compared the risk of adverse events from a then newly approved drug, Rofecoxib (Vioxx), to that from an older drug Naproxen. Vioxx was found to, on average and in comparison to Naproxen, reduce the risk of gastrointestinal (GI) events but increase the risk of thrombotic cardiovascular (CVT) events. Applying StaDISC, we fit 18 popular conditional average treatment effect (CATE) estimators for both outcomes and use calibration to demonstrate their poor global performance. However, they are locally well-calibrated and stable, enabling the identification of patient groups with larger than (estimated) average treatment effects. In fact, StaDISC discovers three clinically interpretable subgroups each for the GI outcome (totaling 29.4% of the study size) and the CVT outcome (totaling 11.0%). Complementary analyses of the found subgroups using the 2001-2004 APPROVe study, a separate independently conducted RCT with 2587 patients, provides further supporting evidence for the promise of StaDISC.
Curating a COVID-19 data repository and forecasting county-level death counts in the United States
May 16, 2020
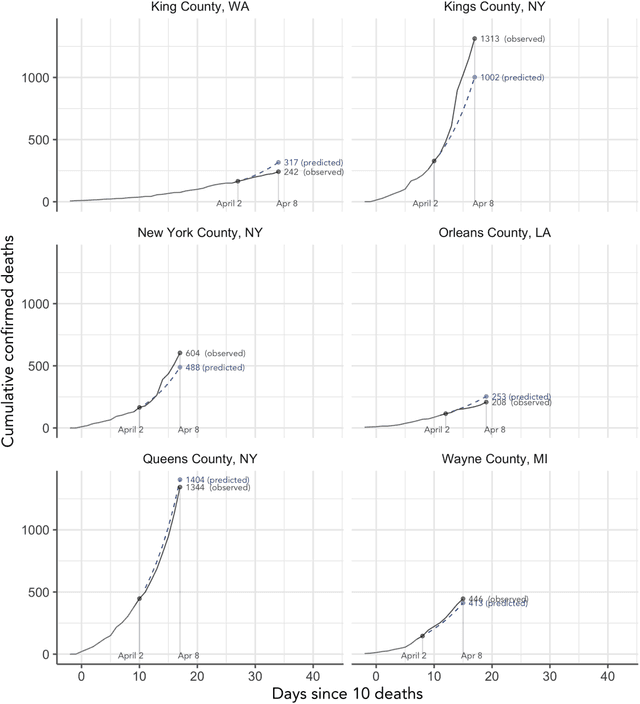
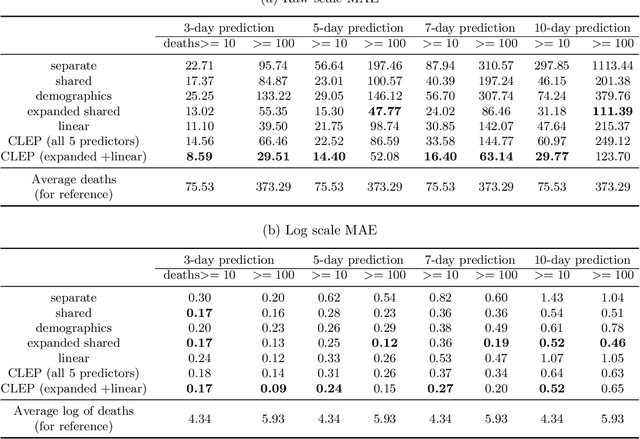
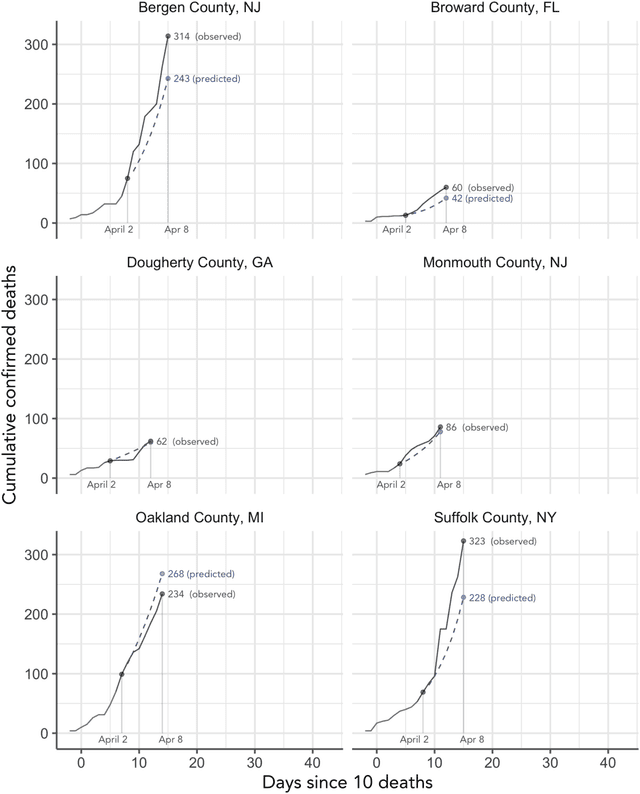
Abstract:As the COVID-19 outbreak continues to evolve, accurate forecasting continues to play an extremely important role in informing policy decisions. In this paper, we collate a large data repository containing COVID-19 information from a range of different sources. We use this data to develop several predictors and prediction intervals for forecasting the short-term (e.g., over the next week) trajectory of COVID-19-related recorded deaths at the county-level in the United States. Specifically, using data from January 22, 2020, to May 10, 2020, we produce several different predictors and combine their forecasts using ensembling techniques, resulting in an ensemble we refer to as Combined Linear and Exponential Predictors (CLEP). Our individual predictors include county-specific exponential and linear predictors, an exponential predictor that pools data together across counties, and a demographics-based exponential predictor. In addition, we use the largest prediction errors in the past five days to assess the uncertainty of our death predictions, resulting in prediction intervals that we refer to as Maximum (absolute) Error Prediction Intervals (MEPI). We show that MEPI is an effective method in practice with a 94.5\% coverage rate when averaged across counties. Our forecasts are already being used by the non-profit organization, Response4Life, to determine the medical supply need for individual hospitals and have directly contributed to the distribution of medical supplies across the country. We hope that our forecasts and data repository can help guide necessary county-specific decision-making and help counties prepare for their continued fight against COVID-19. All collected data, modeling code, forecasts, and visualizations are updated daily and available at \url{https://github.com/Yu-Group/covid19-severity-prediction}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge