Biswajeet Pradhan
ForCM: Forest Cover Mapping from Multispectral Sentinel-2 Image by Integrating Deep Learning with Object-Based Image Analysis
Dec 29, 2025Abstract:This research proposes "ForCM", a novel approach to forest cover mapping that combines Object-Based Image Analysis (OBIA) with Deep Learning (DL) using multispectral Sentinel-2 imagery. The study explores several DL models, including UNet, UNet++, ResUNet, AttentionUNet, and ResNet50-Segnet, applied to high-resolution Sentinel-2 Level 2A satellite images of the Amazon Rainforest. The datasets comprise three collections: two sets of three-band imagery and one set of four-band imagery. After evaluation, the most effective DL models are individually integrated with the OBIA technique to enhance mapping accuracy. The originality of this work lies in evaluating different deep learning models combined with OBIA and comparing them with traditional OBIA methods. The results show that the proposed ForCM method improves forest cover mapping, achieving overall accuracies of 94.54 percent with ResUNet-OBIA and 95.64 percent with AttentionUNet-OBIA, compared to 92.91 percent using traditional OBIA. This research also demonstrates the potential of free and user-friendly tools such as QGIS for accurate mapping within their limitations, supporting global environmental monitoring and conservation efforts.
RAG/LLM Augmented Switching Driven Polymorphic Metaheuristic Framework
May 20, 2025Abstract:Metaheuristic algorithms are widely used for solving complex optimization problems, yet their effectiveness is often constrained by fixed structures and the need for extensive tuning. The Polymorphic Metaheuristic Framework (PMF) addresses this limitation by introducing a self-adaptive metaheuristic switching mechanism driven by real-time performance feedback and dynamic algorithmic selection. PMF leverages the Polymorphic Metaheuristic Agent (PMA) and the Polymorphic Metaheuristic Selection Agent (PMSA) to dynamically select and transition between metaheuristic algorithms based on key performance indicators, ensuring continuous adaptation. This approach enhances convergence speed, adaptability, and solution quality, outperforming traditional metaheuristics in high-dimensional, dynamic, and multimodal environments. Experimental results on benchmark functions demonstrate that PMF significantly improves optimization efficiency by mitigating stagnation and balancing exploration-exploitation strategies across various problem landscapes. By integrating AI-driven decision-making and self-correcting mechanisms, PMF paves the way for scalable, intelligent, and autonomous optimization frameworks, with promising applications in engineering, logistics, and complex decision-making systems.
Optical Coherence Tomography Angiography-OCTA dataset for the study of Diabetic Retinopathy
Sep 06, 2024Abstract:This study presents a dataset consisting of 268 retinal images from 179 individuals, including 133 left-eye and 135 right-eye images, collected from Natasha Eye Care and Research Institute in Pune, Maharashtra, India. The images were captured using a nonmydriatic Optical Coherence Tomography Angiography (OCTA) device, specifically the Optovue Avanti Edition machine as per the protocol mentioned in this paper. Two ophthalmologists then annotated the images. This dataset can be used by researchers and doctors to develop automated diagnostic tools for early detection of diabetic retinopathy (DR).
Full-resolution Lung Nodule Segmentation from Chest X-ray Images using Residual Encoder-Decoder Networks
Jul 13, 2023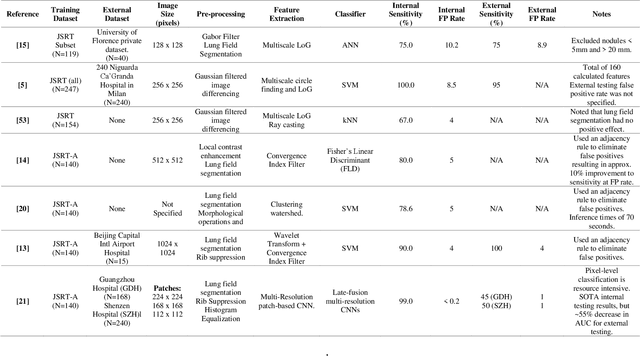
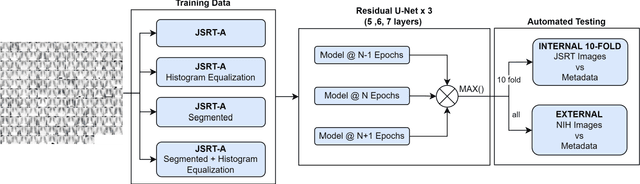
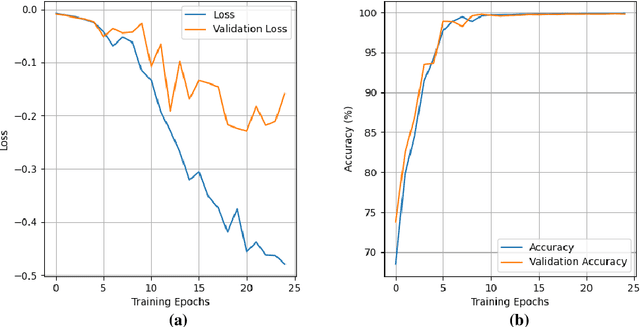

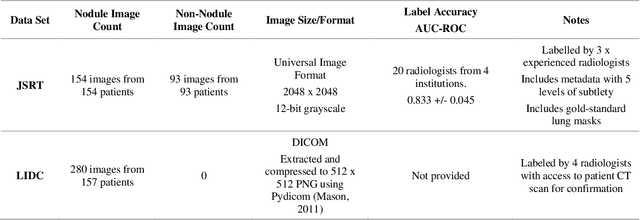
Abstract:Lung cancer is the leading cause of cancer death and early diagnosis is associated with a positive prognosis. Chest X-ray (CXR) provides an inexpensive imaging mode for lung cancer diagnosis. Suspicious nodules are difficult to distinguish from vascular and bone structures using CXR. Computer vision has previously been proposed to assist human radiologists in this task, however, leading studies use down-sampled images and computationally expensive methods with unproven generalization. Instead, this study localizes lung nodules using efficient encoder-decoder neural networks that process full resolution images to avoid any signal loss resulting from down-sampling. Encoder-decoder networks are trained and tested using the JSRT lung nodule dataset. The networks are used to localize lung nodules from an independent external CXR dataset. Sensitivity and false positive rates are measured using an automated framework to eliminate any observer subjectivity. These experiments allow for the determination of the optimal network depth, image resolution and pre-processing pipeline for generalized lung nodule localization. We find that nodule localization is influenced by subtlety, with more subtle nodules being detected in earlier training epochs. Therefore, we propose a novel self-ensemble model from three consecutive epochs centered on the validation optimum. This ensemble achieved a sensitivity of 85% in 10-fold internal testing with false positives of 8 per image. A sensitivity of 81% is achieved at a false positive rate of 6 following morphological false positive reduction. This result is comparable to more computationally complex systems based on linear and spatial filtering, but with a sub-second inference time that is faster than other methods. The proposed algorithm achieved excellent generalization results against an external dataset with sensitivity of 77% at a false positive rate of 7.6.
2-speed network ensemble for efficient classification of incremental land-use/land-cover satellite image chips
Mar 15, 2022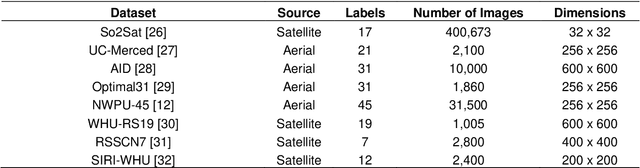
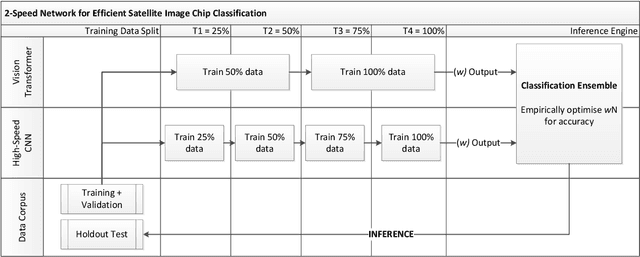

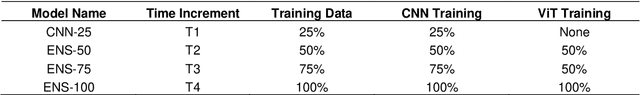
Abstract:The ever-growing volume of satellite imagery data presents a challenge for industry and governments making data-driven decisions based on the timely analysis of very large data sets. Commonly used deep learning algorithms for automatic classification of satellite images are time and resource-intensive to train. The cost of retraining in the context of Big Data presents a practical challenge when new image data and/or classes are added to a training corpus. Recognizing the need for an adaptable, accurate, and scalable satellite image chip classification scheme, in this research we present an ensemble of: i) a slow to train but high accuracy vision transformer; and ii) a fast to train, low-parameter convolutional neural network. The vision transformer model provides a scalable and accurate foundation model. The high-speed CNN provides an efficient means of incorporating newly labelled data into analysis, at the expense of lower accuracy. To simulate incremental data, the very large (~400,000 images) So2Sat LCZ42 satellite image chip dataset is divided into four intervals, with the high-speed CNN retrained every interval and the vision transformer trained every half interval. This experimental setup mimics an increase in data volume and diversity over time. For the task of automated land-cover/land-use classification, the ensemble models for each data increment outperform each of the component models, with best accuracy of 65% against a holdout test partition of the So2Sat dataset. The proposed ensemble and staggered training schedule provide a scalable and cost-effective satellite image classification scheme that is optimized to process very large volumes of satellite data.
Debiasing pipeline improves deep learning model generalization for X-ray based lung nodule detection
Jan 24, 2022

Abstract:Lung cancer is the leading cause of cancer death worldwide and a good prognosis depends on early diagnosis. Unfortunately, screening programs for the early diagnosis of lung cancer are uncommon. This is in-part due to the at-risk groups being located in rural areas far from medical facilities. Reaching these populations would require a scaled approach that combines mobility, low cost, speed, accuracy, and privacy. We can resolve these issues by combining the chest X-ray imaging mode with a federated deep-learning approach, provided that the federated model is trained on homogenous data to ensure that no single data source can adversely bias the model at any point in time. In this study we show that an image pre-processing pipeline that homogenizes and debiases chest X-ray images can improve both internal classification and external generalization, paving the way for a low-cost and accessible deep learning-based clinical system for lung cancer screening. An evolutionary pruning mechanism is used to train a nodule detection deep learning model on the most informative images from a publicly available lung nodule X-ray dataset. Histogram equalization is used to remove systematic differences in image brightness and contrast. Model training is performed using all combinations of lung field segmentation, close cropping, and rib suppression operators. We show that this pre-processing pipeline results in deep learning models that successfully generalize an independent lung nodule dataset using ablation studies to assess the contribution of each operator in this pipeline. In stripping chest X-ray images of known confounding variables by lung field segmentation, along with suppression of signal noise from the bone structure we can train a highly accurate deep learning lung nodule detection algorithm with outstanding generalization accuracy of 89% to nodule samples in unseen data.
Automatic Generation of Interpretable Lung Cancer Scoring Models from Chest X-Ray Images
Dec 17, 2020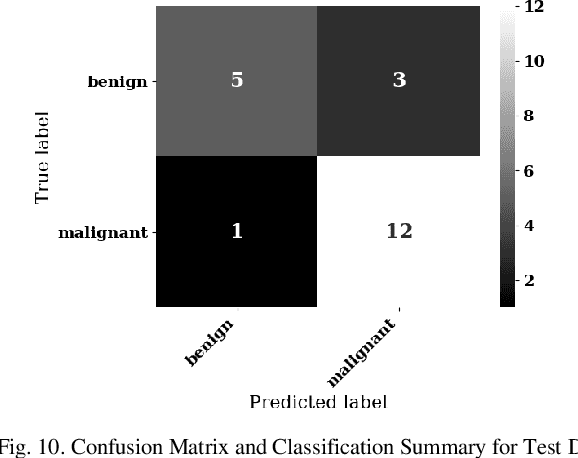
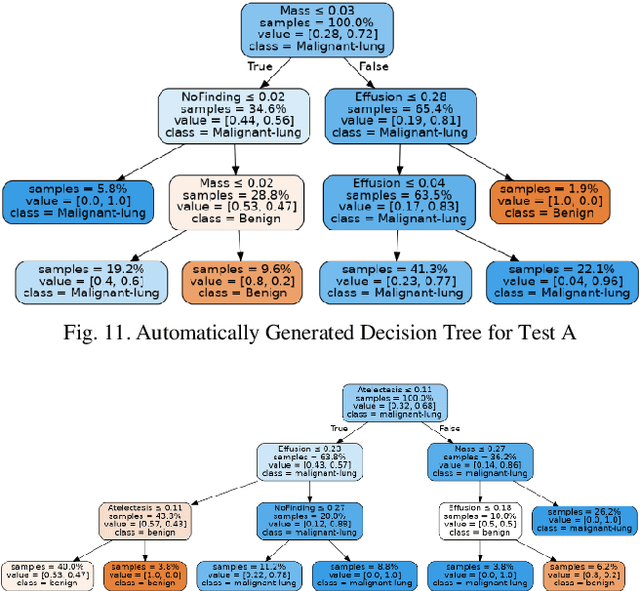
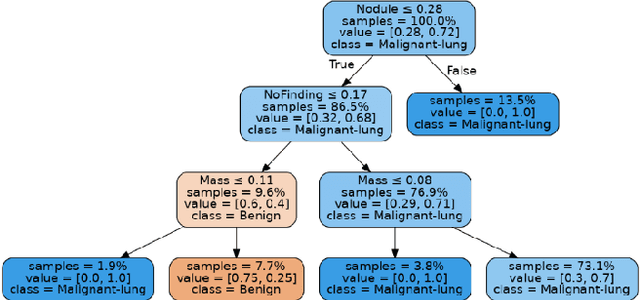
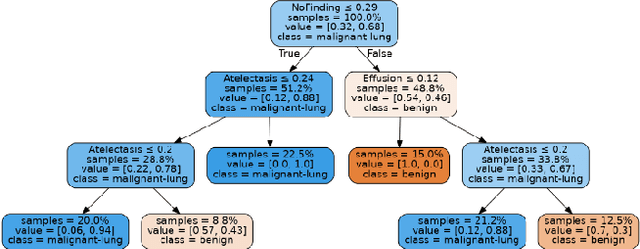
Abstract:Lung cancer is the leading cause of cancer death worldwide with early detection being the key to a positive patient prognosis. Although a multitude of studies have demonstrated that machine learning, and particularly deep learning, techniques are effective at automatically diagnosing lung cancer, these techniques have yet to be clinically approved and adopted by the medical community. Most research in this field is focused on the narrow task of nodule detection to provide an artificial radiological second reading. We instead focus on extracting, from chest X-ray images, a wider range of pathologies associated with lung cancer using a computer vision model trained on a large dataset. We then find the set of best fit decision trees against an independent, smaller dataset for which lung cancer malignancy metadata is provided. For this small inferencing dataset, our best model achieves sensitivity and specificity of 85% and 75% respectively with a positive predictive value of 85% which is comparable to the performance of human radiologists. Furthermore, the decision trees created by this method may be considered as a starting point for refinement by medical experts into clinically usable multi-variate lung cancer scoring and diagnostic models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge