Douglas P. S. Gomes
Improved Abdominal Multi-Organ Segmentation via 3D Boundary-Constrained Deep Neural Networks
Oct 09, 2022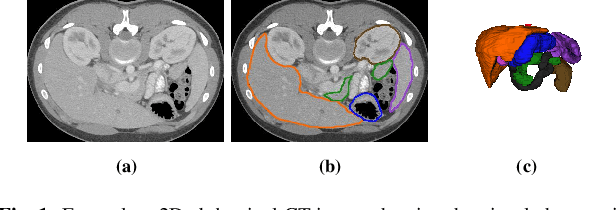
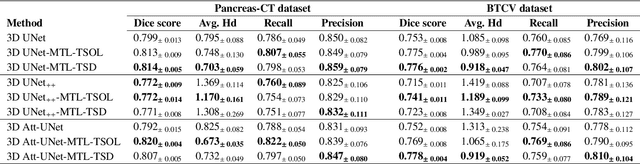
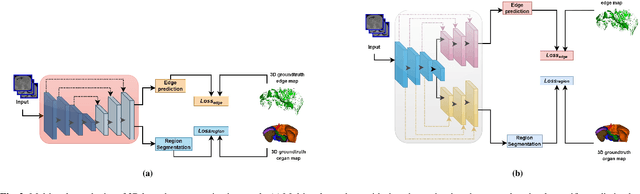
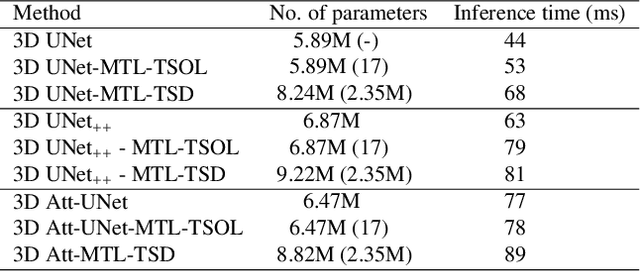
Abstract:Quantitative assessment of the abdominal region from clinically acquired CT scans requires the simultaneous segmentation of abdominal organs. Thanks to the availability of high-performance computational resources, deep learning-based methods have resulted in state-of-the-art performance for the segmentation of 3D abdominal CT scans. However, the complex characterization of organs with fuzzy boundaries prevents the deep learning methods from accurately segmenting these anatomical organs. Specifically, the voxels on the boundary of organs are more vulnerable to misprediction due to the highly-varying intensity of inter-organ boundaries. This paper investigates the possibility of improving the abdominal image segmentation performance of the existing 3D encoder-decoder networks by leveraging organ-boundary prediction as a complementary task. To address the problem of abdominal multi-organ segmentation, we train the 3D encoder-decoder network to simultaneously segment the abdominal organs and their corresponding boundaries in CT scans via multi-task learning. The network is trained end-to-end using a loss function that combines two task-specific losses, i.e., complete organ segmentation loss and boundary prediction loss. We explore two different network topologies based on the extent of weights shared between the two tasks within a unified multi-task framework. To evaluate the utilization of complementary boundary prediction task in improving the abdominal multi-organ segmentation, we use three state-of-the-art encoder-decoder networks: 3D UNet, 3D UNet++, and 3D Attention-UNet. The effectiveness of utilizing the organs' boundary information for abdominal multi-organ segmentation is evaluated on two publically available abdominal CT datasets. A maximum relative improvement of 3.5% and 3.6% is observed in Mean Dice Score for Pancreas-CT and BTCV datasets, respectively.
VeHIF: An Accessible Vegetation High-Impedance Fault Data Set Format
Dec 07, 2021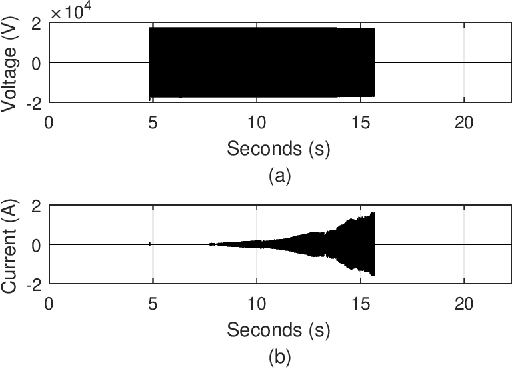
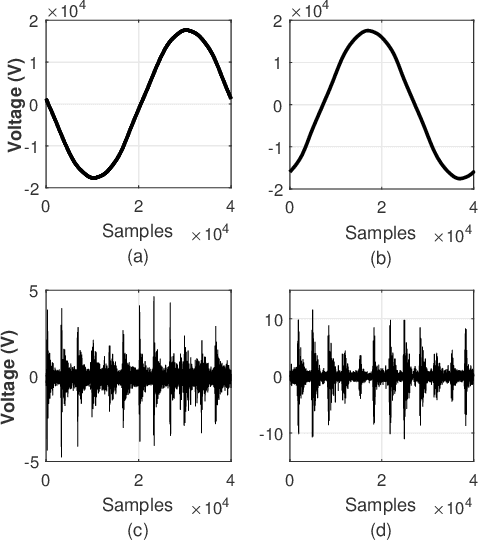
Abstract:High-impedance faults are a challenging problem in power distribution systems. They often do not trigger protection devices and can result in serious hazards such as igniting fires when in contact with vegetation. The current research field dedicated to studying these faults is extensive but suffers from a constraining bottleneck of a lack of real experimental data. Many works set to detect and localize such faults rely on high-impedance fault low-fidelity models, and the lack of public data sets makes it impractical to have objective performance benchmarks. This letter describes and proposes a format for a data set of more than 900 vegetation high-impedance faults funded by the Victorian Government in Australia recorded in high-sampling resolution. The original data set is public, but it was made available through an obscure format that limits its accessibility. The presented format in this letter uses the standard hierarchical data format (HDF5), which makes it easily accessible in many languages such as MATLAB, Python, C++, and more. The data set compiler and visualizer script are also provided in the work repository.
Automatic Generation of Interpretable Lung Cancer Scoring Models from Chest X-Ray Images
Dec 17, 2020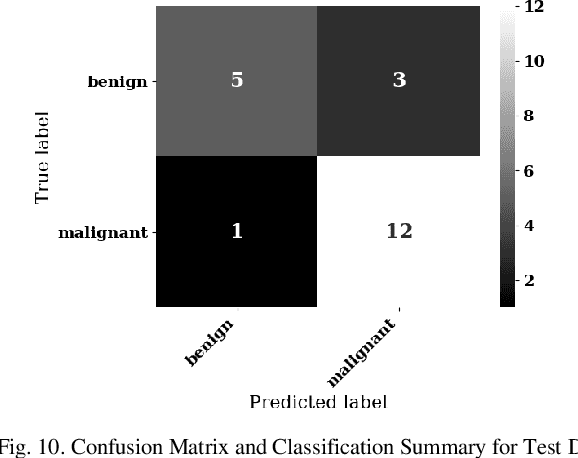
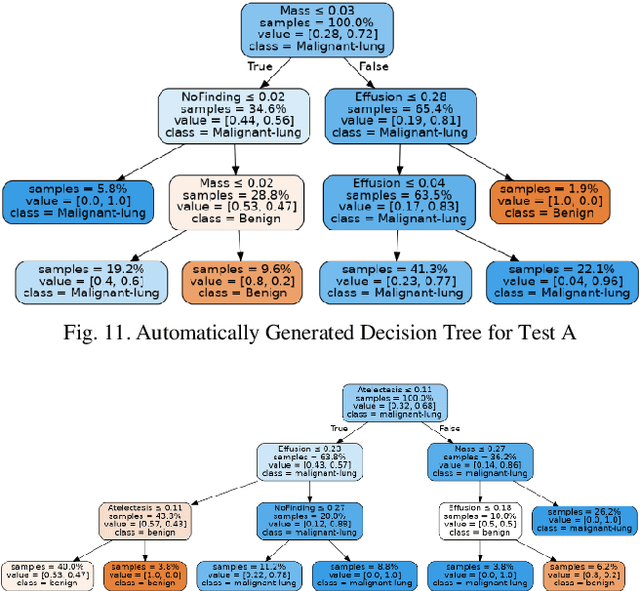
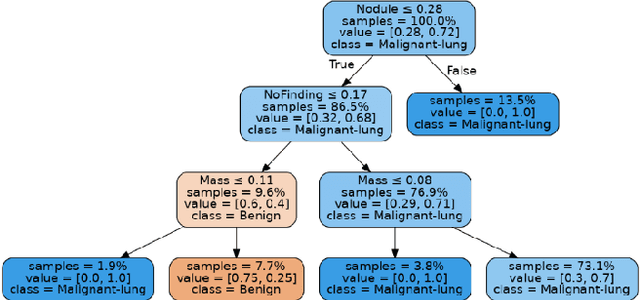
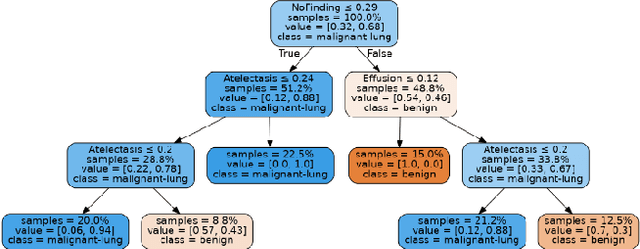
Abstract:Lung cancer is the leading cause of cancer death worldwide with early detection being the key to a positive patient prognosis. Although a multitude of studies have demonstrated that machine learning, and particularly deep learning, techniques are effective at automatically diagnosing lung cancer, these techniques have yet to be clinically approved and adopted by the medical community. Most research in this field is focused on the narrow task of nodule detection to provide an artificial radiological second reading. We instead focus on extracting, from chest X-ray images, a wider range of pathologies associated with lung cancer using a computer vision model trained on a large dataset. We then find the set of best fit decision trees against an independent, smaller dataset for which lung cancer malignancy metadata is provided. For this small inferencing dataset, our best model achieves sensitivity and specificity of 85% and 75% respectively with a positive predictive value of 85% which is comparable to the performance of human radiologists. Furthermore, the decision trees created by this method may be considered as a starting point for refinement by medical experts into clinically usable multi-variate lung cancer scoring and diagnostic models.
MAVIDH Score: A COVID-19 Severity Scoring using Chest X-Ray Pathology Features
Dec 01, 2020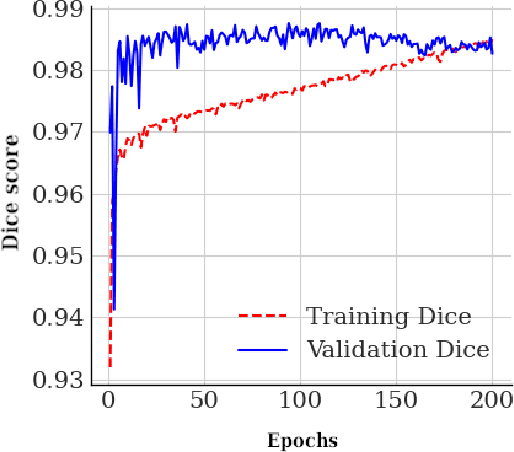
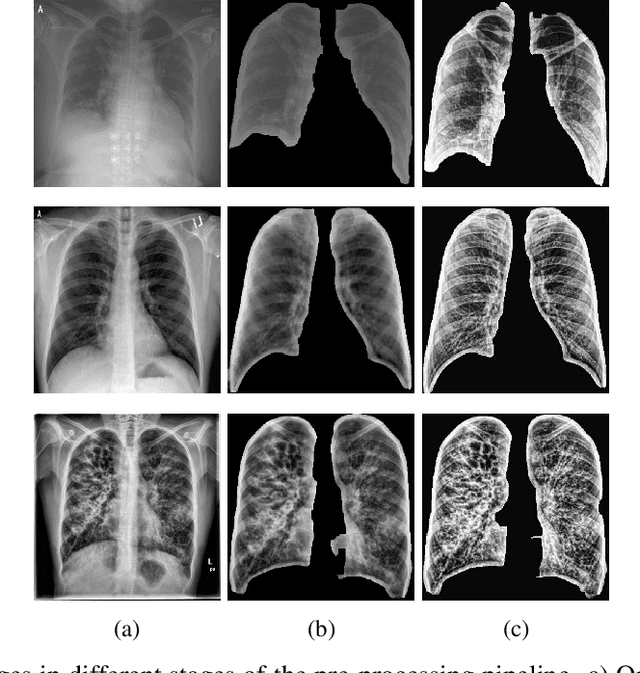
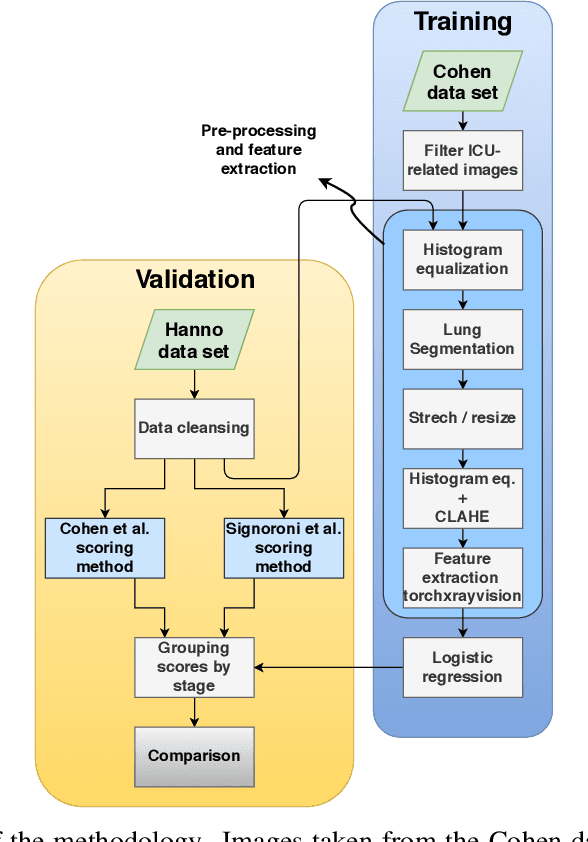
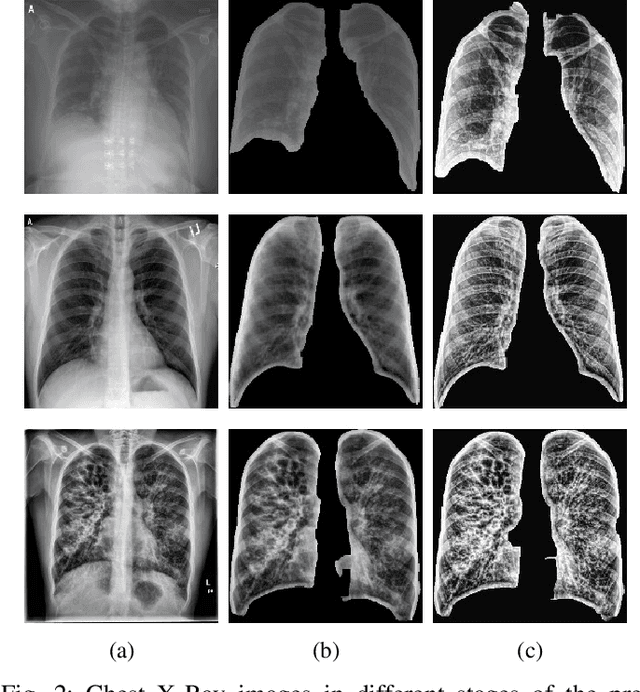
Abstract:The application of computer vision for COVID-19 diagnosis is complex and challenging, given the risks associated with patient misclassifications. Arguably, the primary value of medical imaging for COVID-19 lies rather on patient prognosis. Radiological images can guide physicians assessing the severity of the disease, and a series of images from the same patient at different stages can help to gauge disease progression. Based on these premises, a simple method based on lung-pathology features for scoring disease severity from Chest X-rays is proposed here. As the primary contribution, this method shows to be correlated to patient severity in different stages of disease progression comparatively well when contrasted with other existing methods. An original approach for data selection is also proposed, allowing the simple model to learn the severity-related features. It is hypothesized that the resulting competitive performance presented here is related to the method being feature-based rather than reliant on lung involvement or compromise as others in the literature. The fact that it is simpler and interpretable than other end-to-end, more complex models, also sets aside this work. As the data set is small, bias-inducing artifacts that could lead to overfitting are minimized through an image normalization and lung segmentation step at the learning phase. A second contribution comes from the validation of the results, conceptualized as the scoring of patients groups from different stages of the disease. Besides performing such validation on an independent data set, the results were also compared with other proposed scoring methods in the literature. The expressive results show that although imaging alone is not sufficient for assessing severity as a whole, there is a strong correlation with the scoring system, termed as MAVIDH score, with patient outcome.
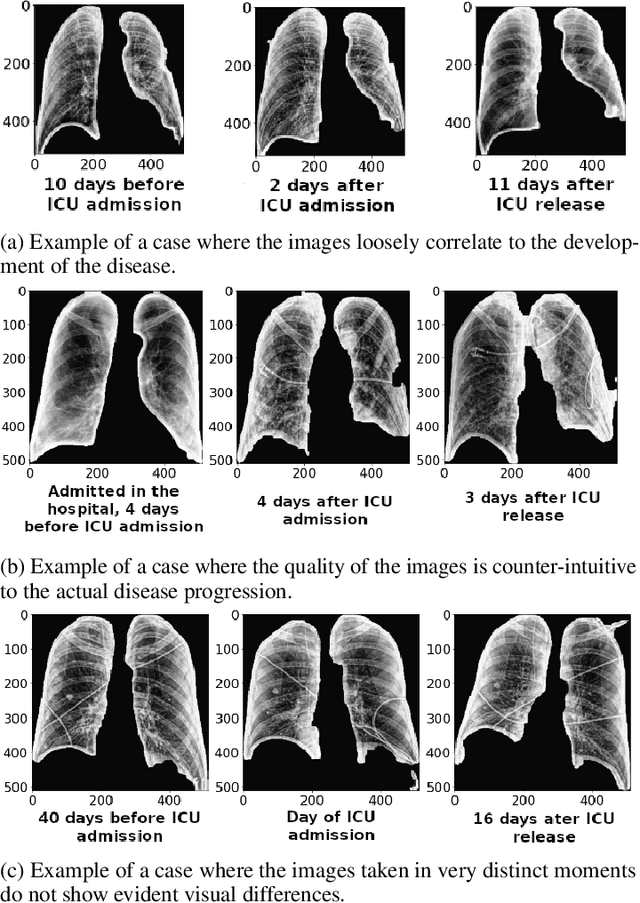
Potential Features of ICU Admission in X-ray Images of COVID-19 Patients
Sep 26, 2020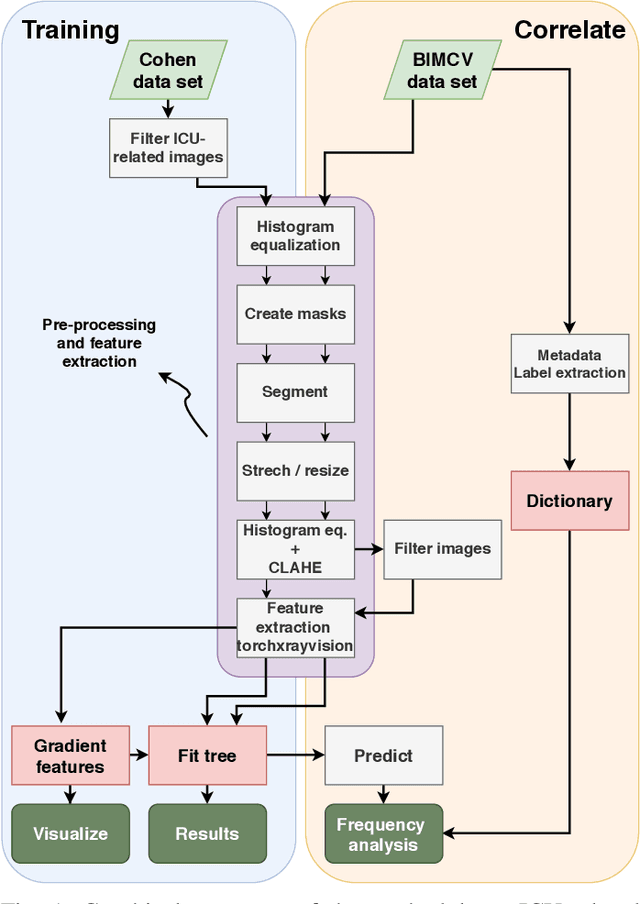
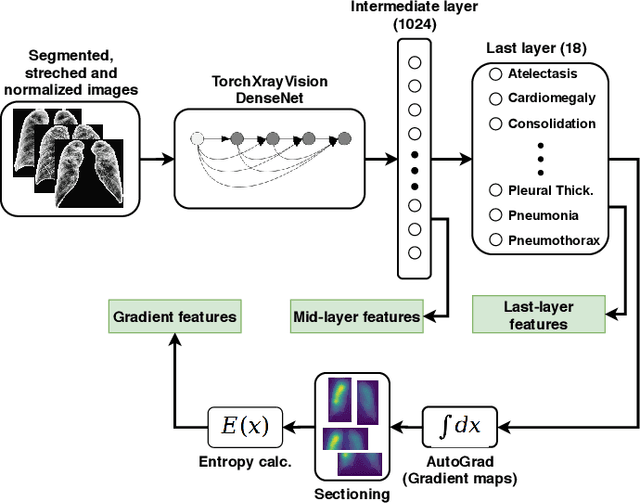

Abstract:X-ray images may present non-trivial features with predictive information of patients that develop severe symptoms of COVID-19. If true, this hypothesis may have practical value in allocating resources to particular patients while using a relatively inexpensive imaging technique. The difficulty of testing such a hypothesis comes from the need for large sets of labelled data, which not only need to be well-annotated but also should contemplate the post-imaging severity outcome. On this account, this paper presents a methodology for extracting features from a limited data set with outcome label (patient required ICU admission or not) and correlating its significance to an additional, larger data set with hundreds of images. The methodology employs a neural network trained to recognise lung pathologies to extract the semantic features, which are then analysed with a shallow decision tree to limit overfitting while increasing interpretability. This analysis points out that only a few features explain most of the variance between patients that developed severe symptoms. When applied to an unrelated, larger data set with labels extracted from clinical notes, the method classified distinct sets of samples where there was a much higher frequency of labels such as `Consolidation', `Effusion', and `alveolar'. A further brief analysis on the locations of such labels also showed a significant increase in the frequency of words like `bilateral', `middle', and `lower' in patients classified as with higher chances of going severe. The methodology for dealing with the lack of specific ICU label data while attesting correlations with a data set containing text notes is novel; its results suggest that some pathologies should receive higher weights when assessing disease severity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge