Baudouin Denis de Senneville
Anatomical feature-prioritized loss for enhanced MR to CT translation
Oct 14, 2024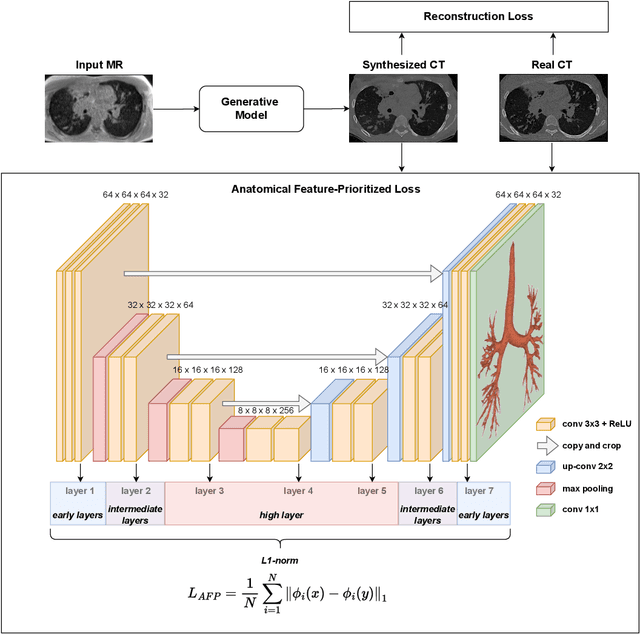
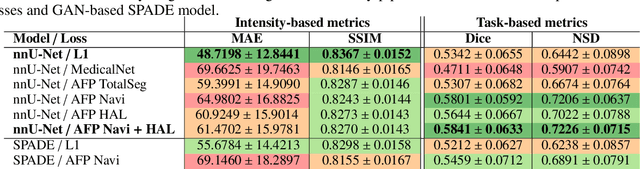
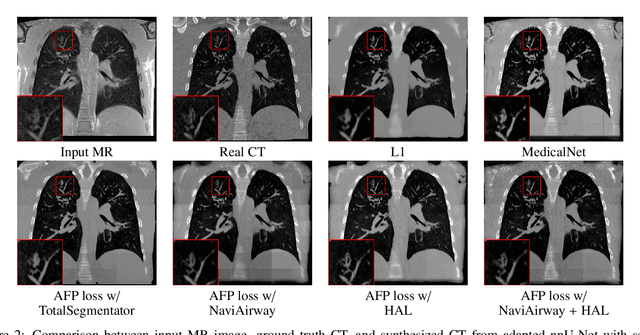
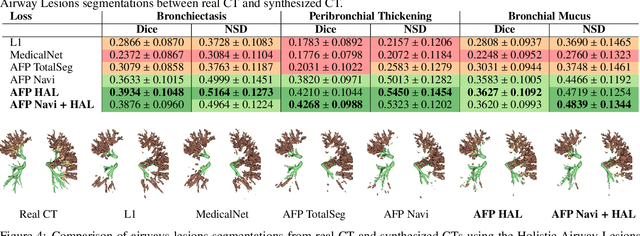
Abstract:In medical image synthesis, the precision of localized structural details is crucial, particularly when addressing specific clinical requirements such as the identification and measurement of fine structures. Traditional methods for image translation and synthesis are generally optimized for global image reconstruction but often fall short in providing the finesse required for detailed local analysis. This study represents a step toward addressing this challenge by introducing a novel anatomical feature-prioritized (AFP) loss function into the synthesis process. This method enhances reconstruction by focusing on clinically significant structures, utilizing features from a pre-trained model designed for a specific downstream task, such as the segmentation of particular anatomical regions. The AFP loss function can replace or complement global reconstruction methods, ensuring a balanced emphasis on both global image fidelity and local structural details. Various implementations of this loss function are explored, including its integration into different synthesis networks such as GAN-based and CNN-based models. Our approach is applied and evaluated in two contexts: lung MR to CT translation, focusing on high-quality reconstruction of bronchial structures, using a private dataset; and pelvis MR to CT synthesis, targeting the accurate representation of organs and muscles, utilizing a public dataset from the Synthrad2023 challenge. We leverage embeddings from pre-trained segmentation models specific to these anatomical regions to demonstrate the capability of the AFP loss to prioritize and accurately reconstruct essential features. This tailored approach shows promising potential for enhancing the specificity and practicality of medical image synthesis in clinical applications.
Enhancing Cell Instance Segmentation in Scanning Electron Microscopy Images via a Deep Contour Closing Operator
Jul 22, 2024Abstract:Accurately segmenting and individualizing cells in SEM images is a highly promising technique for elucidating tissue architecture in oncology. While current AI-based methods are effective, errors persist, necessitating time-consuming manual corrections, particularly in areas where the quality of cell contours in the image is poor and requires gap filling. This study presents a novel AI-driven approach for refining cell boundary delineation to improve instance-based cell segmentation in SEM images, also reducing the necessity for residual manual correction. A CNN COp-Net is introduced to address gaps in cell contours, effectively filling in regions with deficient or absent information. The network takes as input cell contour probability maps with potentially inadequate or missing information and outputs corrected cell contour delineations. The lack of training data was addressed by generating low integrity probability maps using a tailored PDE. We showcase the efficacy of our approach in augmenting cell boundary precision using both private SEM images from PDX hepatoblastoma tissues and publicly accessible images datasets. The proposed cell contour closing operator exhibits a notable improvement in tested datasets, achieving respectively close to 50% (private data) and 10% (public data) increase in the accurately-delineated cell proportion compared to state-of-the-art methods. Additionally, the need for manual corrections was significantly reduced, therefore facilitating the overall digitalization process. Our results demonstrate a notable enhancement in the accuracy of cell instance segmentation, particularly in highly challenging regions where image quality compromises the integrity of cell boundaries, necessitating gap filling. Therefore, our work should ultimately facilitate the study of tumour tissue bioarchitecture in onconanotomy field.
Adaptive local boundary conditions to improve Deformable Image Registration
May 21, 2024Abstract:Objective: In medical imaging, it is often crucial to accurately assess and correct movement during image-guided therapy. Deformable image registration (DIR) consists in estimating the required spatial transformation to align a moving image with a fixed one. However, it is acknowledged that, boundary conditions applied to the solution are critical in preventing mis-registration. Despite the extensive research on registration techniques, relatively few have addressed the issue of boundary conditions in the context of medical DIR. Our aim is a step towards customizing boundary conditions to suit the diverse registration tasks at hand. Approach: We propose a generic, locally adaptive, Robin-type condition enabling to balance between Dirichlet and Neumann boundary conditions, depending on incoming/outgoing flow fields on the image boundaries. The proposed framework is entirely automatized through the determination of a reduced set of hyperparameters optimized via energy minimization. Main results: The proposed approach was tested on a mono-modal CT thorax registration task and an abdominal CT to MRI registration task. For the first task, we observed a relative improvement in terms of target registration error of up to 12% (mean 4%), compared to homogeneous Dirichlet and homogeneous Neumann. For the second task, the automatic framework provides results closed to the best achievable. Significance: This study underscores the importance of tailoring the registration problem at the image boundaries. In this research, we introduce a novel method to adapt the boundary conditions on a voxel-by-voxel basis, yielding optimized results in two distinct tasks: mono-modal CT thorax registration and abdominal CT to MRI registration. The proposed framework enables optimized boundary conditions in image registration without any a priori assumptions regarding the images or the motion.
CT evaluation of 2D and 3D holistic deep learning methods for the volumetric segmentation of airway lesions
Mar 12, 2024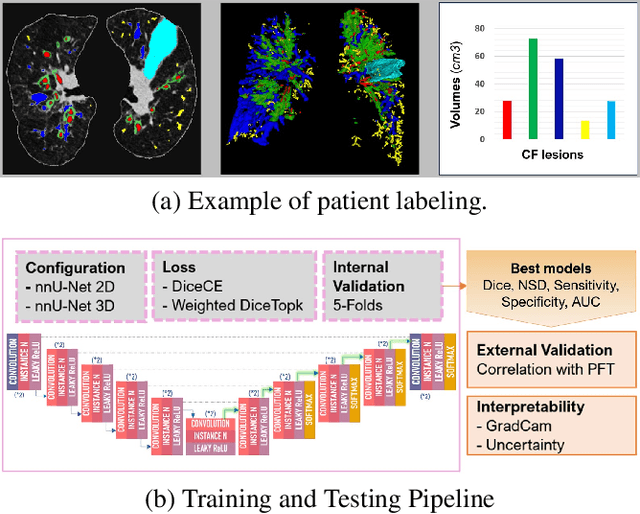
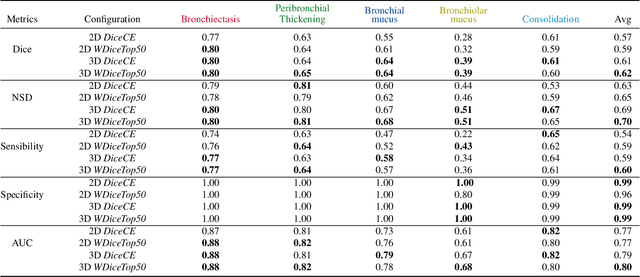
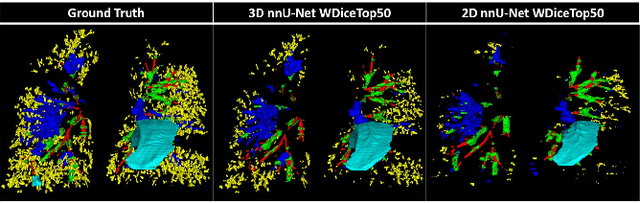
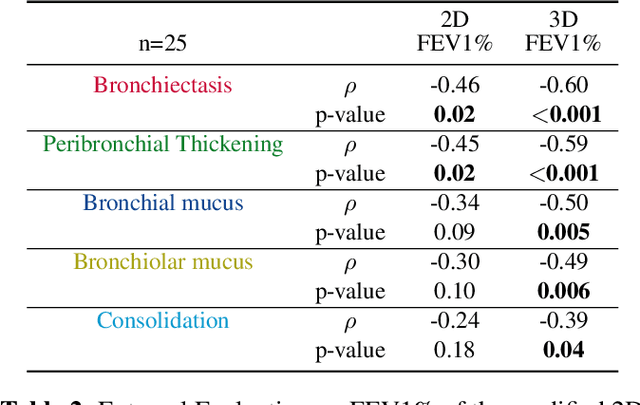
Abstract:This research embarked on a comparative exploration of the holistic segmentation capabilities of Convolutional Neural Networks (CNNs) in both 2D and 3D formats, focusing on cystic fibrosis (CF) lesions. The study utilized data from two CF reference centers, covering five major CF structural changes. Initially, it compared the 2D and 3D models, highlighting the 3D model's superior capability in capturing complex features like mucus plugs and consolidations. To improve the 2D model's performance, a loss adapted to fine structures segmentation was implemented and evaluated, significantly enhancing its accuracy, though not surpassing the 3D model's performance. The models underwent further validation through external evaluation against pulmonary function tests (PFTs), confirming the robustness of the findings. Moreover, this study went beyond comparing metrics; it also included comprehensive assessments of the models' interpretability and reliability, providing valuable insights for their clinical application.
Deep morphological recognition of kidney stones using intra-operative endoscopic digital videos
May 12, 2022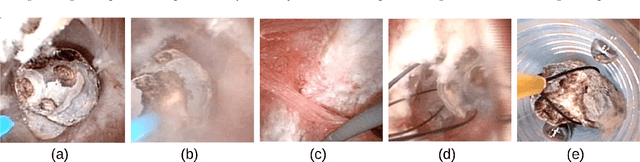
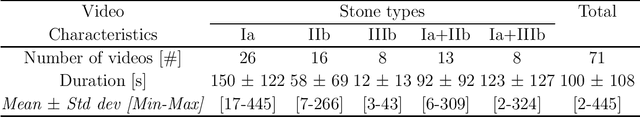
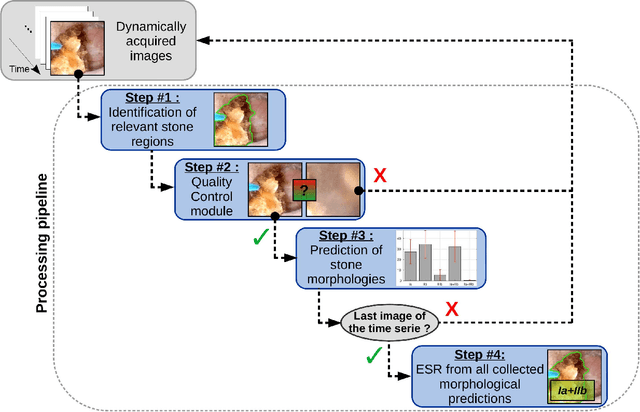
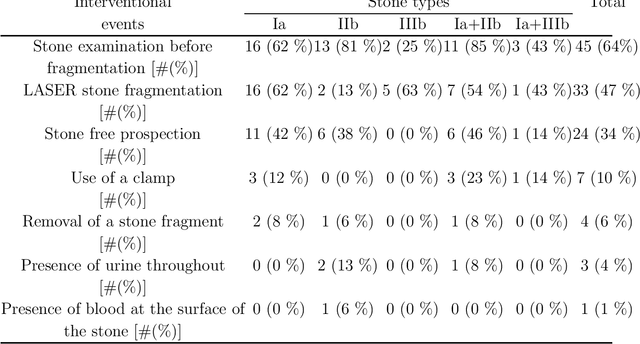
Abstract:The collection and the analysis of kidney stone morphological criteria are essential for an aetiological diagnosis of stone disease. However, in-situ LASER-based fragmentation of urinary stones, which is now the most established chirurgical intervention, may destroy the morphology of the targeted stone. In the current study, we assess the performance and added value of processing complete digital endoscopic video sequences for the automatic recognition of stone morphological features during a standard-of-care intra-operative session. To this end, a computer-aided video classifier was developed to predict in-situ the morphology of stone using an intra-operative digital endoscopic video acquired in a clinical setting. The proposed technique was evaluated on pure (i.e. include one morphology) and mixed (i.e. include at least two morphologies) stones involving "Ia/Calcium Oxalate Monohydrate (COM)", "IIb/ Calcium Oxalate Dihydrate (COD)" and "IIIb/Uric Acid (UA)" morphologies. 71 digital endoscopic videos (50 exhibited only one morphological type and 21 displayed two) were analyzed using the proposed video classifier (56840 frames processed in total). Using the proposed approach, diagnostic performances (averaged over both pure and mixed stone types) were as follows: balanced accuracy=88%, sensitivity=80%, specificity=95%, precision=78% and F1-score=78%. The obtained results demonstrate that AI applied on digital endoscopic video sequences is a promising tool for collecting morphological information during the time-course of the stone fragmentation process without resorting to any human intervention for stone delineation or selection of good quality steady frames. To this end, irrelevant image information must be removed from the prediction process at both frame and pixel levels, which is now feasible thanks to the use of AI-dedicated networks.
Towards Automatic Recognition of Pure & Mixed Stones using Intraoperative Endoscopic Digital Images
May 22, 2021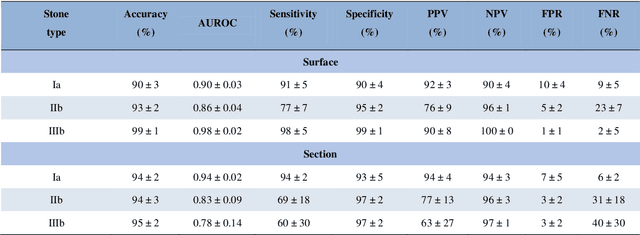
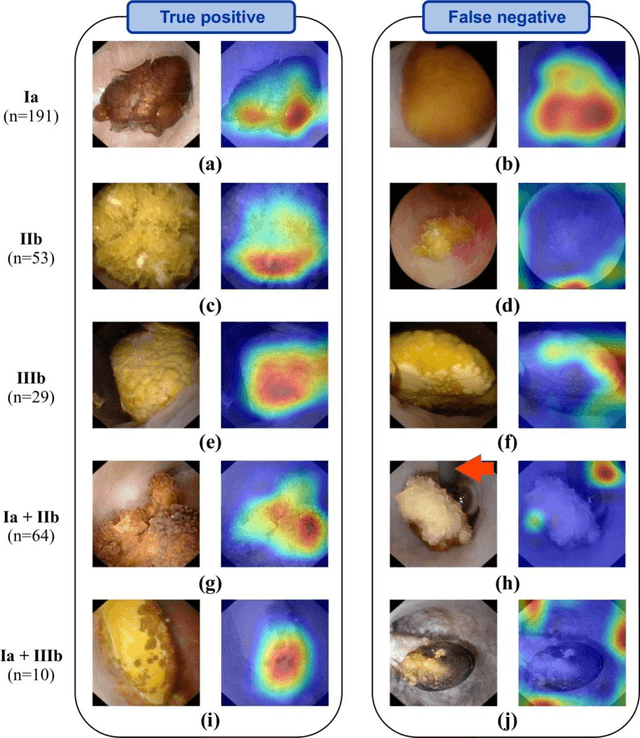
Abstract:Objective: To assess automatic computer-aided in-situ recognition of morphological features of pure and mixed urinary stones using intraoperative digital endoscopic images acquired in a clinical setting. Materials and methods: In this single-centre study, an experienced urologist intraoperatively and prospectively examined the surface and section of all kidney stones encountered. Calcium oxalate monohydrate (COM/Ia), dihydrate (COD/IIb) and uric acid (UA/IIIb) morphological criteria were collected and classified to generate annotated datasets. A deep convolutional neural network (CNN) was trained to predict the composition of both pure and mixed stones. To explain the predictions of the deep neural network model, coarse localisation heat-maps were plotted to pinpoint key areas identified by the network. Results: This study included 347 and 236 observations of stone surface and stone section, respectively. A highest sensitivity of 98 % was obtained for the type "pure IIIb/UA" using surface images. The most frequently encountered morphology was that of the type "pure Ia/COM"; it was correctly predicted in 91 % and 94 % of cases using surface and section images, respectively. Of the mixed type "Ia/COM+IIb/COD", Ia/COM was predicted in 84 % of cases using surface images, IIb/COD in 70 % of cases, and both in 65 % of cases. Concerning mixed Ia/COM+IIIb/UA stones, Ia/COM was predicted in 91 % of cases using section images, IIIb/UA in 69 % of cases, and both in 74 % of cases. Conclusions: This preliminary study demonstrates that deep convolutional neural networks are promising to identify kidney stone composition from endoscopic images acquired intraoperatively. Both pure and mixed stone composition could be discriminated. Collected in a clinical setting, surface and section images analysed by deep CNN provide valuable information about stone morphology for computer-aided diagnosis.
MRI-Guided High Intensity Focused Ultrasound of Liver and Kidney
Nov 21, 2020



Abstract:High Intensity Focused Ultrasound (HIFU) can be used to achieve a local temperature increase deep inside the human body in a non-invasive way. MRI guidance of the procedure allows in situ target definition. In addition, MRI can be used to provide continuous temperature mapping during HIFU for spatial and temporal control of the heating procedure and prediction of the final lesion based on the received thermal dose. Temperature mapping of mobile organs as kidney and liver is challenging, as well as real-time processing methods for feedback control of the HIFU procedure. In this paper, recent technological advances are reviewed in MR temperature mapping of these organs, in motion compensation of the HIFU beam, in intercostal HIFU sonication, and in volumetric ablation and feedback control strategies. Recent pre-clinical studies have demonstrated the feasibility of each of these novel methods. The perspectives to translate those advances into the clinic are addressed. It can be concluded that MR guided HIFU for ablation in liver and kidney appears feasible but requires further work on integration of technologically advanced methods.
Deep correction of breathing-related artifacts in MR-thermometry
Nov 10, 2020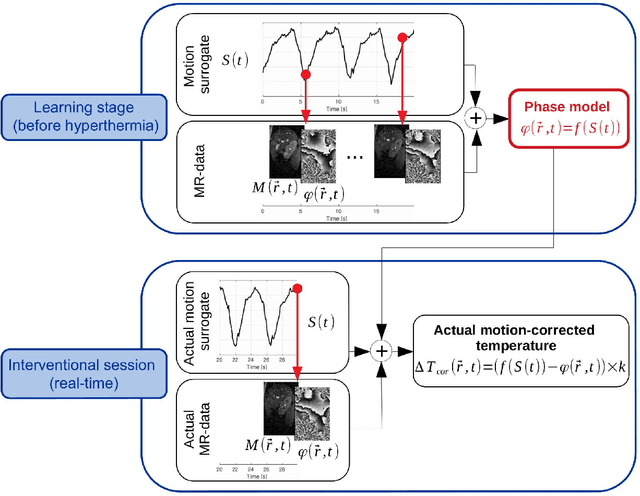
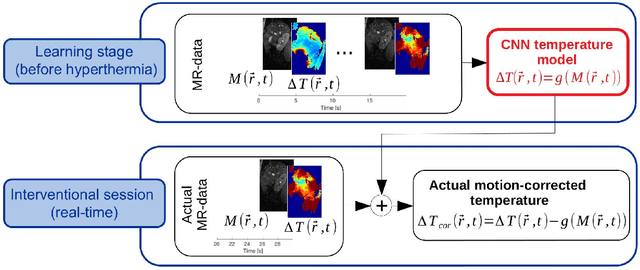
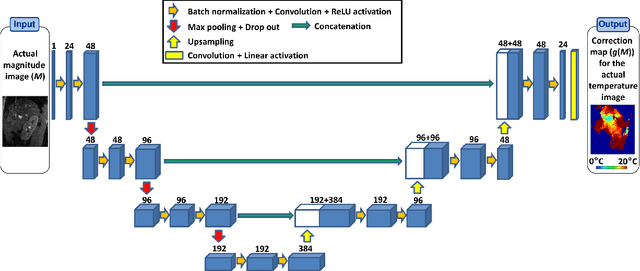
Abstract:Real-time MR-imaging has been clinically adapted for monitoring thermal therapies since it can provide on-the-fly temperature maps simultaneously with anatomical information. However, proton resonance frequency based thermometry of moving targets remains challenging since temperature artifacts are induced by the respiratory as well as physiological motion. If left uncorrected, these artifacts lead to severe errors in temperature estimates and impair therapy guidance. In this study, we evaluated deep learning for on-line correction of motion related errors in abdominal MR-thermometry. For this, a convolutional neural network (CNN) was designed to learn the apparent temperature perturbation from images acquired during a preparative learning stage prior to hyperthermia. The input of the designed CNN is the most recent magnitude image and no surrogate of motion is needed. During the subsequent hyperthermia procedure, the recent magnitude image is used as an input for the CNN-model in order to generate an on-line correction for the current temperature map. The method's artifact suppression performance was evaluated on 12 free breathing volunteers and was found robust and artifact-free in all examined cases. Furthermore, thermometric precision and accuracy was assessed for in vivo ablation using high intensity focused ultrasound. All calculations involved at the different stages of the proposed workflow were designed to be compatible with the clinical time constraints of a therapeutic procedure.
RegQCNET: Deep Quality Control for Image-to-template Brain MRI Registration
May 14, 2020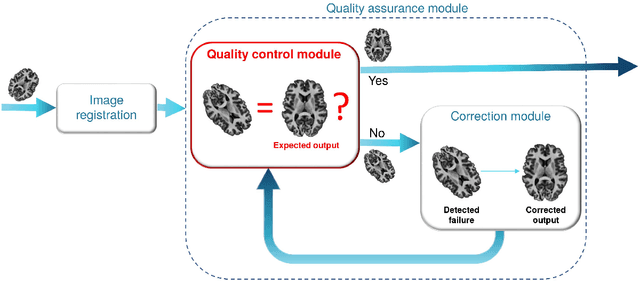
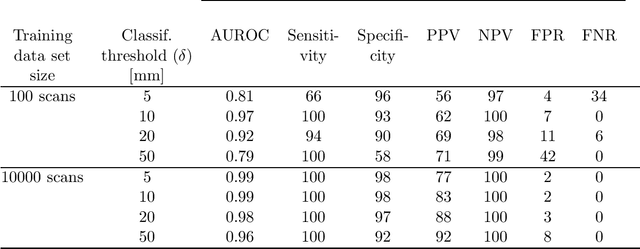
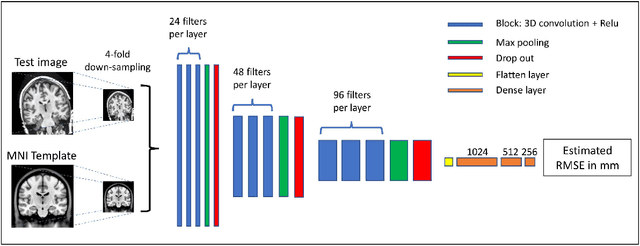
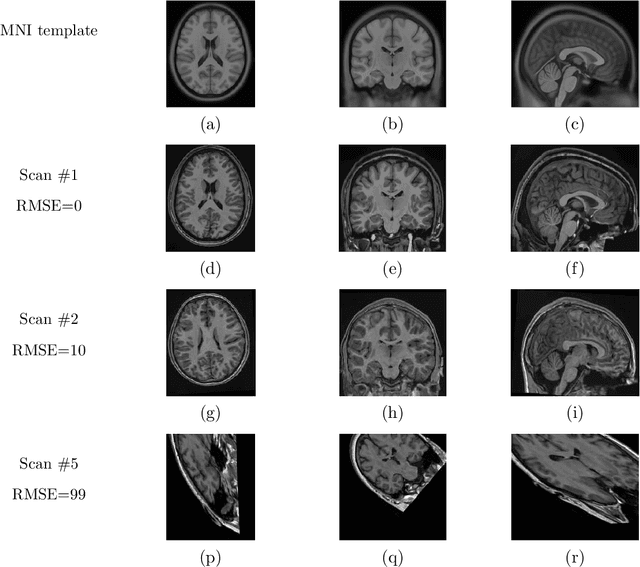
Abstract:Registration of one or several brain image(s) onto a common reference space defined by a template is a necessary prerequisite for many image processing tasks, such as brain structure segmentation or functional MRI study. Manual assessment of registration quality is a tedious and time-consuming task, especially when a large amount of data is involved. An automated and reliable quality control (QC) is thus mandatory. Moreover, the computation time of the QC must be also compatible with the processing of massive datasets. Therefore, deep neural network approaches appear as a method of choice to automatically assess registration quality. In the current study, a compact 3D CNN, referred to as RegQCNET, is introduced to quantitatively predict the amplitude of a registration mismatch between the registered image and the reference template. This quantitative estimation of registration error is expressed using metric unit system. Therefore, a meaningful task-specific threshold can be manually or automatically defined in order to distinguish usable and non-usable images. The robustness of the proposed RegQCNET is first analyzed on lifespan brain images undergoing various simulated spatial transformations and intensity variations between training and testing. Secondly, the potential of RegQCNET to classify images as usable or non-usable is evaluated using both manual and automatic thresholds. The latters were estimated using several computer-assisted classification models through cross-validation. To this end we used expert's visual quality control estimated on a lifespan cohort of 3953 brains. Finally, the RegQCNET accuracy is compared to usual image features such as image correlation coefficient and mutual information. Results show that the proposed deep learning QC is robust, fast and accurate to estimate registration error in processing pipeline.
AssemblyNet: A large ensemble of CNNs for 3D Whole Brain MRI Segmentation
Nov 20, 2019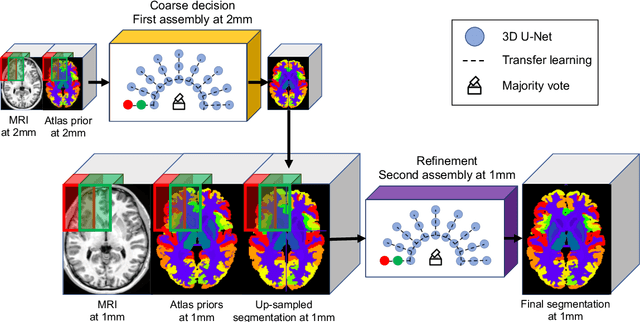
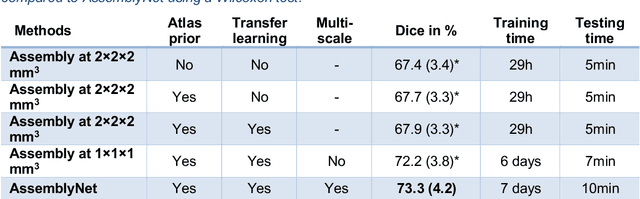
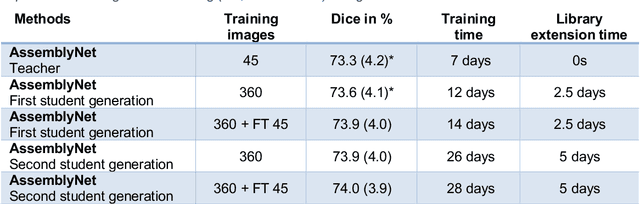
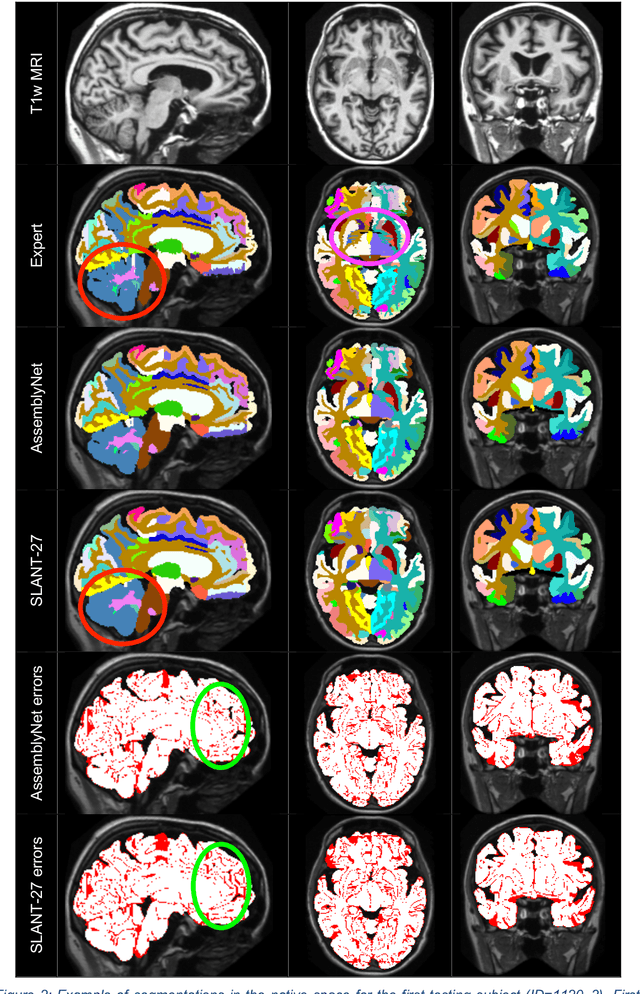
Abstract:Whole brain segmentation using deep learning (DL) is a very challenging task since the number of anatomical labels is very high compared to the number of available training images. To address this problem, previous DL methods proposed to use a single convolution neural network (CNN) or few independent CNNs. In this paper, we present a novel ensemble method based on a large number of CNNs processing different overlapping brain areas. Inspired by parliamentary decision-making systems, we propose a framework called AssemblyNet, made of two "assemblies" of U-Nets. Such a parliamentary system is capable of dealing with complex decisions, unseen problem and reaching a consensus quickly. AssemblyNet introduces sharing of knowledge among neighboring U-Nets, an "amendment" procedure made by the second assembly at higher-resolution to refine the decision taken by the first one, and a final decision obtained by majority voting. During our validation, AssemblyNet showed competitive performance compared to state-of-the-art methods such as U-Net, Joint label fusion and SLANT. Moreover, we investigated the scan-rescan consistency and the robustness to disease effects of our method. These experiences demonstrated the reliability of AssemblyNet. Finally, we showed the interest of using semi-supervised learning to improve the performance of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge