Ba-Hien Tran
Huawei Paris Research Center
Universal Adaptive Environment Discovery
Oct 14, 2025Abstract:An open problem in Machine Learning is how to avoid models to exploit spurious correlations in the data; a famous example is the background-label shortcut in the Waterbirds dataset. A common remedy is to train a model across multiple environments; in the Waterbirds dataset, this corresponds to training by randomizing the background. However, selecting the right environments is a challenging problem, given that these are rarely known a priori. We propose Universal Adaptive Environment Discovery (UAED), a unified framework that learns a distribution over data transformations that instantiate environments, and optimizes any robust objective averaged over this learned distribution. UAED yields adaptive variants of IRM, REx, GroupDRO, and CORAL without predefined groups or manual environment design. We provide a theoretical analysis by providing PAC-Bayes bounds and by showing robustness to test environment distributions under standard conditions. Empirically, UAED discovers interpretable environment distributions and improves worst-case accuracy on standard benchmarks, while remaining competitive on mean accuracy. Our results indicate that making environments adaptive is a practical route to out-of-distribution generalization.
Highly Efficient and Effective LLMs with Multi-Boolean Architectures
May 28, 2025



Abstract:Weight binarization has emerged as a promising strategy to drastically reduce the complexity of large language models (LLMs). It is mainly classified into two approaches: post-training binarization and finetuning with training-aware binarization methods. The first approach, while having low complexity, leads to significant loss of information from the original LLMs, resulting in poor performance. The second approach, on the other hand, relies heavily on full-precision latent weights for gradient approximation of binary weights, which not only remains suboptimal but also introduces substantial complexity. In this paper, we introduce a novel framework that effectively transforms LLMs into multi-kernel Boolean parameters, for the first time, finetunes them directly in the Boolean domain, eliminating the need for expensive latent weights. This significantly reduces complexity during both finetuning and inference. Through extensive and insightful experiments across a wide range of LLMs, we demonstrate that our method outperforms recent ultra low-bit quantization and binarization methods.
Optimizing Data Augmentation through Bayesian Model Selection
May 27, 2025Abstract:Data Augmentation (DA) has become an essential tool to improve robustness and generalization of modern machine learning. However, when deciding on DA strategies it is critical to choose parameters carefully, and this can be a daunting task which is traditionally left to trial-and-error or expensive optimization based on validation performance. In this paper, we counter these limitations by proposing a novel framework for optimizing DA. In particular, we take a probabilistic view of DA, which leads to the interpretation of augmentation parameters as model (hyper)-parameters, and the optimization of the marginal likelihood with respect to these parameters as a Bayesian model selection problem. Due to its intractability, we derive a tractable Evidence Lower BOund (ELBO), which allows us to optimize augmentation parameters jointly with model parameters. We provide extensive theoretical results on variational approximation quality, generalization guarantees, invariance properties, and connections to empirical Bayes. Through experiments on computer vision tasks, we show that our approach improves calibration and yields robust performance over fixed or no augmentation. Our work provides a rigorous foundation for optimizing DA through Bayesian principles with significant potential for robust machine learning.
Robust Classification by Coupling Data Mollification with Label Smoothing
Jun 03, 2024



Abstract:Introducing training-time augmentations is a key technique to enhance generalization and prepare deep neural networks against test-time corruptions. Inspired by the success of generative diffusion models, we propose a novel approach coupling data augmentation, in the form of image noising and blurring, with label smoothing to align predicted label confidences with image degradation. The method is simple to implement, introduces negligible overheads, and can be combined with existing augmentations. We demonstrate improved robustness and uncertainty quantification on the corrupted image benchmarks of the CIFAR and TinyImageNet datasets.
BOLD: Boolean Logic Deep Learning
May 25, 2024Abstract:Deep learning is computationally intensive, with significant efforts focused on reducing arithmetic complexity, particularly regarding energy consumption dominated by data movement. While existing literature emphasizes inference, training is considerably more resource-intensive. This paper proposes a novel mathematical principle by introducing the notion of Boolean variation such that neurons made of Boolean weights and inputs can be trained -- for the first time -- efficiently in Boolean domain using Boolean logic instead of gradient descent and real arithmetic. We explore its convergence, conduct extensively experimental benchmarking, and provide consistent complexity evaluation by considering chip architecture, memory hierarchy, dataflow, and arithmetic precision. Our approach achieves baseline full-precision accuracy in ImageNet classification and surpasses state-of-the-art results in semantic segmentation, with notable performance in image super-resolution, and natural language understanding with transformer-based models. Moreover, it significantly reduces energy consumption during both training and inference.
Spatial Bayesian Neural Networks
Nov 16, 2023Abstract:Statistical models for spatial processes play a central role in statistical analyses of spatial data. Yet, it is the simple, interpretable, and well understood models that are routinely employed even though, as is revealed through prior and posterior predictive checks, these can poorly characterise the spatial heterogeneity in the underlying process of interest. Here, we propose a new, flexible class of spatial-process models, which we refer to as spatial Bayesian neural networks (SBNNs). An SBNN leverages the representational capacity of a Bayesian neural network; it is tailored to a spatial setting by incorporating a spatial "embedding layer" into the network and, possibly, spatially-varying network parameters. An SBNN is calibrated by matching its finite-dimensional distribution at locations on a fine gridding of space to that of a target process of interest. That process could be easy to simulate from or we have many realisations from it. We propose several variants of SBNNs, most of which are able to match the finite-dimensional distribution of the target process at the selected grid better than conventional BNNs of similar complexity. We also show that a single SBNN can be used to represent a variety of spatial processes often used in practice, such as Gaussian processes and lognormal processes. We briefly discuss the tools that could be used to make inference with SBNNs, and we conclude with a discussion of their advantages and limitations.
One-Line-of-Code Data Mollification Improves Optimization of Likelihood-based Generative Models
May 30, 2023



Abstract:Generative Models (GMs) have attracted considerable attention due to their tremendous success in various domains, such as computer vision where they are capable to generate impressive realistic-looking images. Likelihood-based GMs are attractive due to the possibility to generate new data by a single model evaluation. However, they typically achieve lower sample quality compared to state-of-the-art score-based diffusion models (DMs). This paper provides a significant step in the direction of addressing this limitation. The idea is to borrow one of the strengths of score-based DMs, which is the ability to perform accurate density estimation in low-density regions and to address manifold overfitting by means of data mollification. We connect data mollification through the addition of Gaussian noise to Gaussian homotopy, which is a well-known technique to improve optimization. Data mollification can be implemented by adding one line of code in the optimization loop, and we demonstrate that this provides a boost in generation quality of likelihood-based GMs, without computational overheads. We report results on image data sets with popular likelihood-based GMs, including variants of variational autoencoders and normalizing flows, showing large improvements in FID score.
Fully Bayesian Autoencoders with Latent Sparse Gaussian Processes
Feb 09, 2023



Abstract:Autoencoders and their variants are among the most widely used models in representation learning and generative modeling. However, autoencoder-based models usually assume that the learned representations are i.i.d. and fail to capture the correlations between the data samples. To address this issue, we propose a novel Sparse Gaussian Process Bayesian Autoencoder (SGPBAE) model in which we impose fully Bayesian sparse Gaussian Process priors on the latent space of a Bayesian Autoencoder. We perform posterior estimation for this model via stochastic gradient Hamiltonian Monte Carlo. We evaluate our approach qualitatively and quantitatively on a wide range of representation learning and generative modeling tasks and show that our approach consistently outperforms multiple alternatives relying on Variational Autoencoders.
Model Selection for Bayesian Autoencoders
Jun 11, 2021
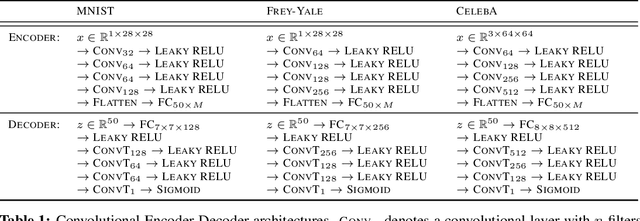
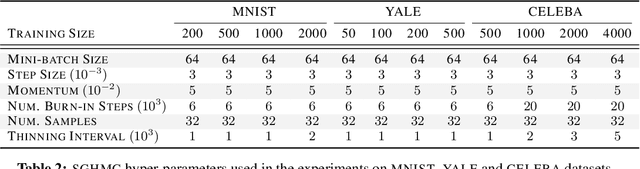

Abstract:We develop a novel method for carrying out model selection for Bayesian autoencoders (BAEs) by means of prior hyper-parameter optimization. Inspired by the common practice of type-II maximum likelihood optimization and its equivalence to Kullback-Leibler divergence minimization, we propose to optimize the distributional sliced-Wasserstein distance (DSWD) between the output of the autoencoder and the empirical data distribution. The advantages of this formulation are that we can estimate the DSWD based on samples and handle high-dimensional problems. We carry out posterior estimation of the BAE parameters via stochastic gradient Hamiltonian Monte Carlo and turn our BAE into a generative model by fitting a flexible Dirichlet mixture model in the latent space. Consequently, we obtain a powerful alternative to variational autoencoders, which are the preferred choice in modern applications of autoencoders for representation learning with uncertainty. We evaluate our approach qualitatively and quantitatively using a vast experimental campaign on a number of unsupervised learning tasks and show that, in small-data regimes where priors matter, our approach provides state-of-the-art results, outperforming multiple competitive baselines.
All You Need is a Good Functional Prior for Bayesian Deep Learning
Nov 25, 2020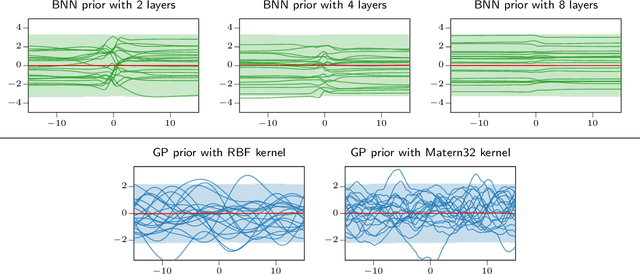
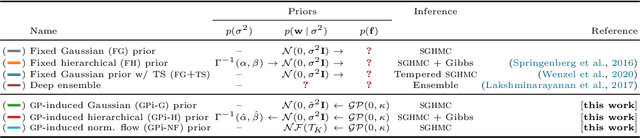

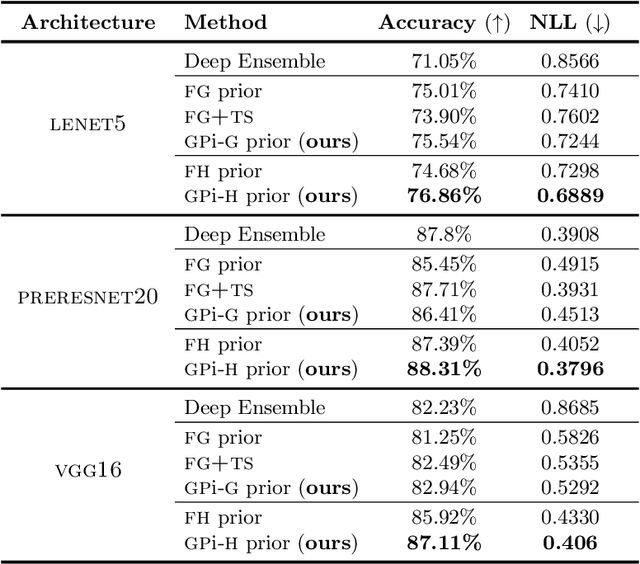
Abstract:The Bayesian treatment of neural networks dictates that a prior distribution is specified over their weight and bias parameters. This poses a challenge because modern neural networks are characterized by a large number of parameters, and the choice of these priors has an uncontrolled effect on the induced functional prior, which is the distribution of the functions obtained by sampling the parameters from their prior distribution. We argue that this is a hugely limiting aspect of Bayesian deep learning, and this work tackles this limitation in a practical and effective way. Our proposal is to reason in terms of functional priors, which are easier to elicit, and to "tune" the priors of neural network parameters in a way that they reflect such functional priors. Gaussian processes offer a rigorous framework to define prior distributions over functions, and we propose a novel and robust framework to match their prior with the functional prior of neural networks based on the minimization of their Wasserstein distance. We provide vast experimental evidence that coupling these priors with scalable Markov chain Monte Carlo sampling offers systematically large performance improvements over alternative choices of priors and state-of-the-art approximate Bayesian deep learning approaches. We consider this work a considerable step in the direction of making the long-standing challenge of carrying out a fully Bayesian treatment of neural networks, including convolutional neural networks, a concrete possibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge