Auguste Genovesio
XFACTORS: Disentangled Information Bottleneck via Contrastive Supervision
Jan 29, 2026Abstract:Disentangled representation learning aims to map independent factors of variation to independent representation components. On one hand, purely unsupervised approaches have proven successful on fully disentangled synthetic data, but fail to recover semantic factors from real data without strong inductive biases. On the other hand, supervised approaches are unstable and hard to scale to large attribute sets because they rely on adversarial objectives or auxiliary classifiers. We introduce \textsc{XFactors}, a weakly-supervised VAE framework that disentangles and provides explicit control over a chosen set of factors. Building on the Disentangled Information Bottleneck perspective, we decompose the representation into a residual subspace $\mathcal{S}$ and factor-specific subspaces $\mathcal{T}_1,\ldots,\mathcal{T}_K$ and a residual subspace $\mathcal{S}$. Each target factor is encoded in its assigned $\mathcal{T}_i$ through contrastive supervision: an InfoNCE loss pulls together latents sharing the same factor value and pushes apart mismatched pairs. In parallel, KL regularization imposes a Gaussian structure on both $\mathcal{S}$ and the aggregated factor subspaces, organizing the geometry without additional supervision for non-targeted factors and avoiding adversarial training and classifiers. Across multiple datasets, with constant hyperparameters, \textsc{XFactors} achieves state-of-the-art disentanglement scores and yields consistent qualitative factor alignment in the corresponding subspaces, enabling controlled factor swapping via latent replacement. We further demonstrate that our method scales correctly with increasing latent capacity and evaluate it on the real-world dataset CelebA. Our code is available at \href{https://github.com/ICML26-anon/XFactors}{github.com/ICML26-anon/XFactors}.
Multi-marginal temporal Schrödinger Bridge Matching for video generation from unpaired data
Oct 02, 2025Abstract:Many natural dynamic processes -- such as in vivo cellular differentiation or disease progression -- can only be observed through the lens of static sample snapshots. While challenging, reconstructing their temporal evolution to decipher underlying dynamic properties is of major interest to scientific research. Existing approaches enable data transport along a temporal axis but are poorly scalable in high dimension and require restrictive assumptions to be met. To address these issues, we propose \textit{\textbf{Multi-Marginal temporal Schr\"odinger Bridge Matching}} (\textbf{MMtSBM}) \textit{for video generation from unpaired data}, extending the theoretical guarantees and empirical efficiency of Diffusion Schr\"odinger Bridge Matching (arXiv:archive/2303.16852) by deriving the Iterative Markovian Fitting algorithm to multiple marginals in a novel factorized fashion. Experiments show that MMtSBM retains theoretical properties on toy examples, achieves state-of-the-art performance on real world datasets such as transcriptomic trajectory inference in 100 dimensions, and for the first time recovers couplings and dynamics in very high dimensional image settings. Our work establishes multi-marginal Schr\"odinger bridges as a practical and principled approach for recovering hidden dynamics from static data.
A Cross Modal Knowledge Distillation & Data Augmentation Recipe for Improving Transcriptomics Representations through Morphological Features
May 27, 2025Abstract:Understanding cellular responses to stimuli is crucial for biological discovery and drug development. Transcriptomics provides interpretable, gene-level insights, while microscopy imaging offers rich predictive features but is harder to interpret. Weakly paired datasets, where samples share biological states, enable multimodal learning but are scarce, limiting their utility for training and multimodal inference. We propose a framework to enhance transcriptomics by distilling knowledge from microscopy images. Using weakly paired data, our method aligns and binds modalities, enriching gene expression representations with morphological information. To address data scarcity, we introduce (1) Semi-Clipped, an adaptation of CLIP for cross-modal distillation using pretrained foundation models, achieving state-of-the-art results, and (2) PEA (Perturbation Embedding Augmentation), a novel augmentation technique that enhances transcriptomics data while preserving inherent biological information. These strategies improve the predictive power and retain the interpretability of transcriptomics, enabling rich unimodal representations for complex biological tasks.
GANs Conditioning Methods: A Survey
Sep 03, 2024Abstract:In recent years, Generative Adversarial Networks (GANs) have seen significant advancements, leading to their widespread adoption across various fields. The original GAN architecture enables the generation of images without any specific control over the content, making it an unconditional generation process. However, many practical applications require precise control over the generated output, which has led to the development of conditional GANs (cGANs) that incorporate explicit conditioning to guide the generation process. cGANs extend the original framework by incorporating additional information (conditions), enabling the generation of samples that adhere to that specific criteria. Various conditioning methods have been proposed, each differing in how they integrate the conditioning information into both the generator and the discriminator networks. In this work, we review the conditioning methods proposed for GANs, exploring the characteristics of each method and highlighting their unique mechanisms and theoretical foundations. Furthermore, we conduct a comparative analysis of these methods, evaluating their performance on various image datasets. Through these analyses, we aim to provide insights into the strengths and limitations of various conditioning techniques, guiding future research and application in generative modeling.
Reconstructing Interpretable Features in Computational Super-Resolution microscopy via Regularized Latent Search
May 29, 2024Abstract:Supervised deep learning approaches can artificially increase the resolution of microscopy images by learning a mapping between two image resolutions or modalities. However, such methods often require a large set of hard-to-get low-res/high-res image pairs and produce synthetic images with a moderate increase in resolution. Conversely, recent methods based on GAN latent search offered a drastic increase in resolution without the need of paired images. However, they offer limited reconstruction of the high-resolution image interpretable features. Here, we propose a robust super-resolution method based on regularized latent search~(RLS) that offers an actionable balance between fidelity to the ground-truth and realism of the recovered image given a distribution prior. The latter allows to split the analysis of a low-resolution image into a computational super-resolution task performed by deep learning followed by a quantification task performed by a handcrafted algorithm and based on interpretable biological features. This two-step process holds potential for various applications such as diagnostics on mobile devices, where the main aim is not to recover the high-resolution details of a specific sample but rather to obtain high-resolution images that preserve explainable and quantifiable differences between conditions.
PhenDiff: Revealing Invisible Phenotypes with Conditional Diffusion Models
Dec 13, 2023Abstract:Over the last five years, deep generative models have gradually been adopted for various tasks in biological research. Notably, image-to-image translation methods showed to be effective in revealing subtle phenotypic cell variations otherwise invisible to the human eye. Current methods to achieve this goal mainly rely on Generative Adversarial Networks (GANs). However, these models are known to suffer from some shortcomings such as training instability and mode collapse. Furthermore, the lack of robustness to invert a real image into the latent of a trained GAN prevents flexible editing of real images. In this work, we propose PhenDiff, an image-to-image translation method based on conditional diffusion models to identify subtle phenotypes in microscopy images. We evaluate this approach on biological datasets against previous work such as CycleGAN. We show that PhenDiff outperforms this baseline in terms of quality and diversity of the generated images. We then apply this method to display invisible phenotypic changes triggered by a rare neurodevelopmental disorder on microscopy images of organoids. Altogether, we demonstrate that PhenDiff is able to perform high quality biological image-to-image translation allowing to spot subtle phenotype variations on a real image.
Super-Resolution through StyleGAN Regularized Latent Search: A Realism-Fidelity Trade-off
Nov 28, 2023Abstract:This paper addresses the problem of super-resolution: constructing a highly resolved (HR) image from a low resolved (LR) one. Recent unsupervised approaches search the latent space of a StyleGAN pre-trained on HR images, for the image that best downscales to the input LR image. However, they tend to produce out-of-domain images and fail to accurately reconstruct HR images that are far from the original domain. Our contribution is twofold. Firstly, we introduce a new regularizer to constrain the search in the latent space, ensuring that the inverted code lies in the original image manifold. Secondly, we further enhanced the reconstruction through expanding the image prior around the optimal latent code. Our results show that the proposed approach recovers realistic high-quality images for large magnification factors. Furthermore, for low magnification factors, it can still reconstruct details that the generator could not have produced otherwise. Altogether, our approach achieves a good trade-off between fidelity and realism for the super-resolution task.
ChAda-ViT : Channel Adaptive Attention for Joint Representation Learning of Heterogeneous Microscopy Images
Nov 26, 2023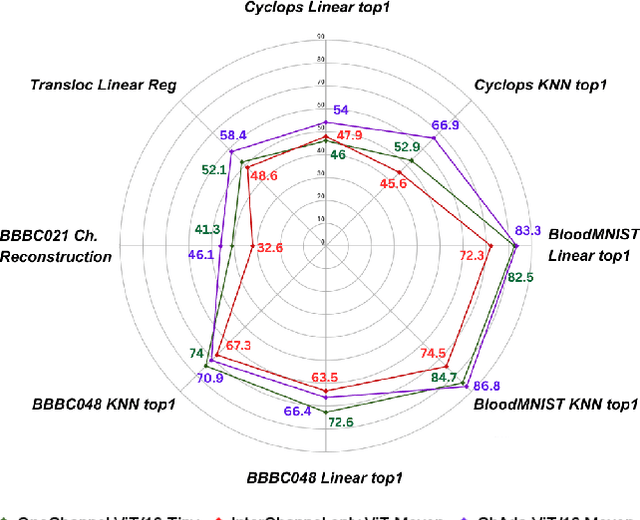
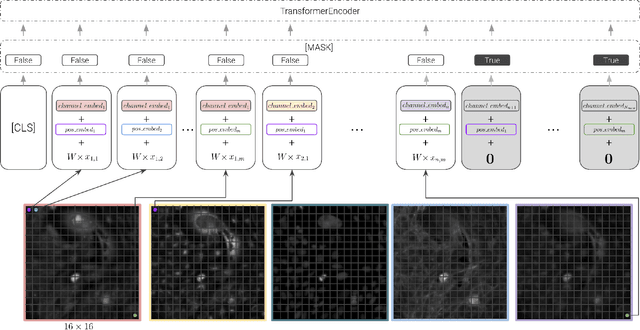

Abstract:Unlike color photography images, which are consistently encoded into RGB channels, biological images encompass various modalities, where the type of microscopy and the meaning of each channel varies with each experiment. Importantly, the number of channels can range from one to a dozen and their correlation is often comparatively much lower than RGB, as each of them brings specific information content. This aspect is largely overlooked by methods designed out of the bioimage field, and current solutions mostly focus on intra-channel spatial attention, often ignoring the relationship between channels, yet crucial in most biological applications. Importantly, the variable channel type and count prevent the projection of several experiments to a unified representation for large scale pre-training. In this study, we propose ChAda-ViT, a novel Channel Adaptive Vision Transformer architecture employing an Inter-Channel Attention mechanism on images with an arbitrary number, order and type of channels. We also introduce IDRCell100k, a bioimage dataset with a rich set of 79 experiments covering 7 microscope modalities, with a multitude of channel types, and channel counts varying from 1 to 10 per experiment. Our proposed architecture, trained in a self-supervised manner, outperforms existing approaches in several biologically relevant downstream tasks. Additionally, it can be used to bridge the gap for the first time between assays with different microscopes, channel numbers or types by embedding various image and experimental modalities into a unified biological image representation. The latter should facilitate interdisciplinary studies and pave the way for better adoption of deep learning in biological image-based analyses. Code and Data to be released soon.
One Style is All you Need to Generate a Video
Oct 27, 2023Abstract:In this paper, we propose a style-based conditional video generative model. We introduce a novel temporal generator based on a set of learned sinusoidal bases. Our method learns dynamic representations of various actions that are independent of image content and can be transferred between different actors. Beyond the significant enhancement of video quality compared to prevalent methods, we demonstrate that the disentangled dynamic and content permit their independent manipulation, as well as temporal GAN-inversion to retrieve and transfer a video motion from one content or identity to another without further preprocessing such as landmark points.
No Free Lunch in Self Supervised Representation Learning
Apr 23, 2023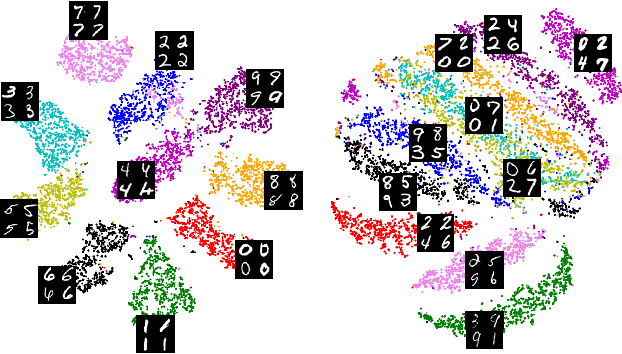

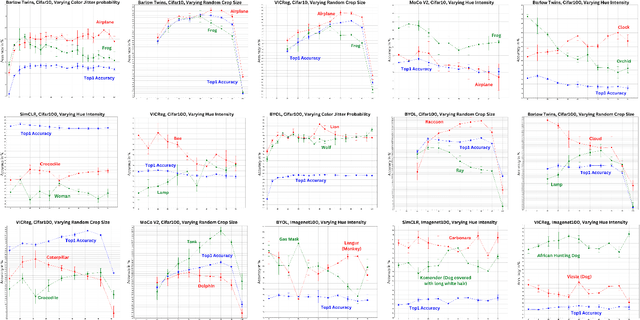
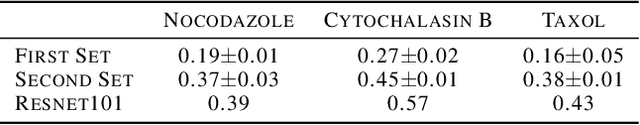
Abstract:Self-supervised representation learning in computer vision relies heavily on hand-crafted image transformations to learn meaningful and invariant features. However few extensive explorations of the impact of transformation design have been conducted in the literature. In particular, the dependence of downstream performances to transformation design has been established, but not studied in depth. In this work, we explore this relationship, its impact on a domain other than natural images, and show that designing the transformations can be viewed as a form of supervision. First, we demonstrate that not only do transformations have an effect on downstream performance and relevance of clustering, but also that each category in a supervised dataset can be impacted in a different way. Following this, we explore the impact of transformation design on microscopy images, a domain where the difference between classes is more subtle and fuzzy than in natural images. In this case, we observe a greater impact on downstream tasks performances. Finally, we demonstrate that transformation design can be leveraged as a form of supervision, as careful selection of these by a domain expert can lead to a drastic increase in performance on a given downstream task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge