Ethan Cohen
ChAda-ViT : Channel Adaptive Attention for Joint Representation Learning of Heterogeneous Microscopy Images
Nov 26, 2023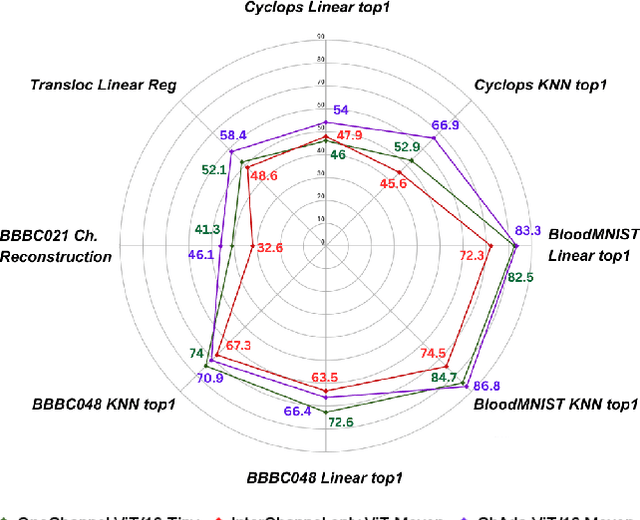
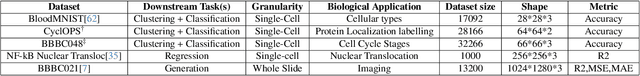
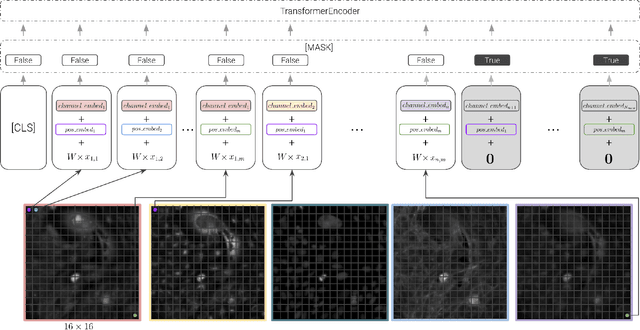

Abstract:Unlike color photography images, which are consistently encoded into RGB channels, biological images encompass various modalities, where the type of microscopy and the meaning of each channel varies with each experiment. Importantly, the number of channels can range from one to a dozen and their correlation is often comparatively much lower than RGB, as each of them brings specific information content. This aspect is largely overlooked by methods designed out of the bioimage field, and current solutions mostly focus on intra-channel spatial attention, often ignoring the relationship between channels, yet crucial in most biological applications. Importantly, the variable channel type and count prevent the projection of several experiments to a unified representation for large scale pre-training. In this study, we propose ChAda-ViT, a novel Channel Adaptive Vision Transformer architecture employing an Inter-Channel Attention mechanism on images with an arbitrary number, order and type of channels. We also introduce IDRCell100k, a bioimage dataset with a rich set of 79 experiments covering 7 microscope modalities, with a multitude of channel types, and channel counts varying from 1 to 10 per experiment. Our proposed architecture, trained in a self-supervised manner, outperforms existing approaches in several biologically relevant downstream tasks. Additionally, it can be used to bridge the gap for the first time between assays with different microscopes, channel numbers or types by embedding various image and experimental modalities into a unified biological image representation. The latter should facilitate interdisciplinary studies and pave the way for better adoption of deep learning in biological image-based analyses. Code and Data to be released soon.
Reconstructing the Mind's Eye: fMRI-to-Image with Contrastive Learning and Diffusion Priors
May 29, 2023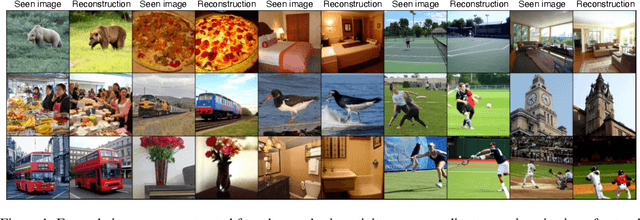
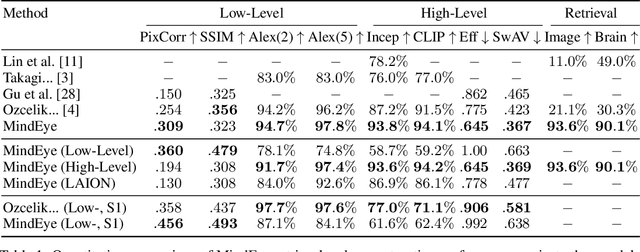
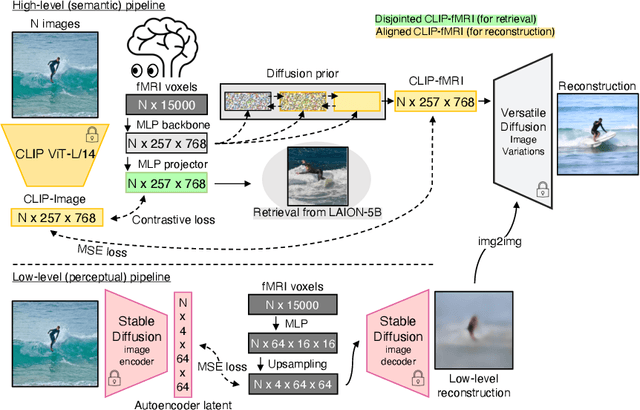
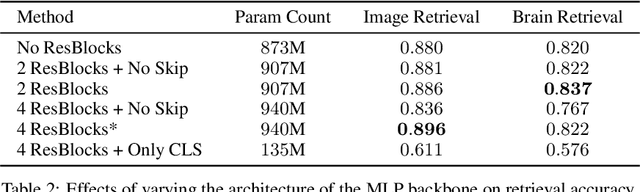
Abstract:We present MindEye, a novel fMRI-to-image approach to retrieve and reconstruct viewed images from brain activity. Our model comprises two parallel submodules that are specialized for retrieval (using contrastive learning) and reconstruction (using a diffusion prior). MindEye can map fMRI brain activity to any high dimensional multimodal latent space, like CLIP image space, enabling image reconstruction using generative models that accept embeddings from this latent space. We comprehensively compare our approach with other existing methods, using both qualitative side-by-side comparisons and quantitative evaluations, and show that MindEye achieves state-of-the-art performance in both reconstruction and retrieval tasks. In particular, MindEye can retrieve the exact original image even among highly similar candidates indicating that its brain embeddings retain fine-grained image-specific information. This allows us to accurately retrieve images even from large-scale databases like LAION-5B. We demonstrate through ablations that MindEye's performance improvements over previous methods result from specialized submodules for retrieval and reconstruction, improved training techniques, and training models with orders of magnitude more parameters. Furthermore, we show that MindEye can better preserve low-level image features in the reconstructions by using img2img, with outputs from a separate autoencoder. All code is available on GitHub.
No Free Lunch in Self Supervised Representation Learning
Apr 23, 2023

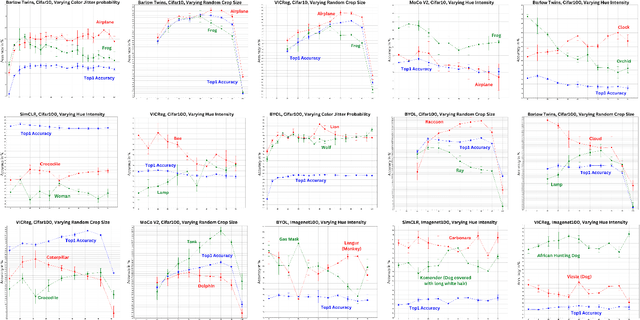
Abstract:Self-supervised representation learning in computer vision relies heavily on hand-crafted image transformations to learn meaningful and invariant features. However few extensive explorations of the impact of transformation design have been conducted in the literature. In particular, the dependence of downstream performances to transformation design has been established, but not studied in depth. In this work, we explore this relationship, its impact on a domain other than natural images, and show that designing the transformations can be viewed as a form of supervision. First, we demonstrate that not only do transformations have an effect on downstream performance and relevance of clustering, but also that each category in a supervised dataset can be impacted in a different way. Following this, we explore the impact of transformation design on microscopy images, a domain where the difference between classes is more subtle and fuzzy than in natural images. In this case, we observe a greater impact on downstream tasks performances. Finally, we demonstrate that transformation design can be leveraged as a form of supervision, as careful selection of these by a domain expert can lead to a drastic increase in performance on a given downstream task.
Comparison of semi-supervised learning methods for High Content Screening quality control
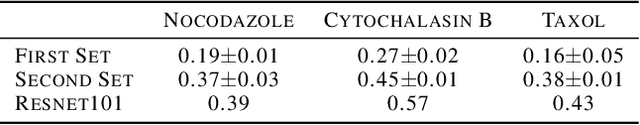
Aug 09, 2022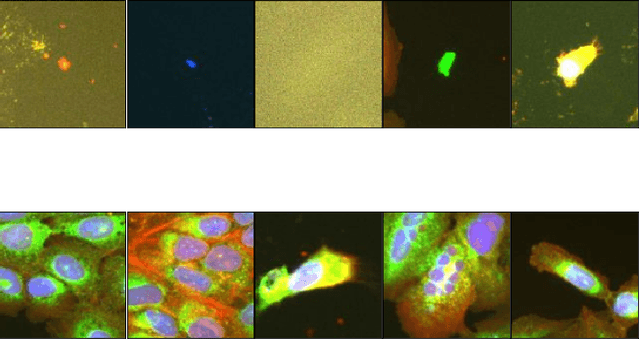

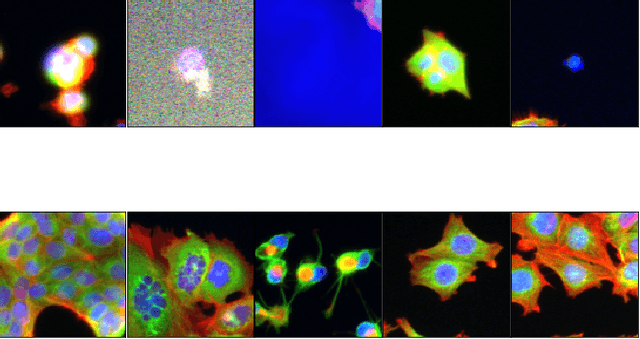

Abstract:Progress in automated microscopy and quantitative image analysis has promoted high-content screening (HCS) as an efficient drug discovery and research tool. While HCS offers to quantify complex cellular phenotypes from images at high throughput, this process can be obstructed by image aberrations such as out-of-focus image blur, fluorophore saturation, debris, a high level of noise, unexpected auto-fluorescence or empty images. While this issue has received moderate attention in the literature, overlooking these artefacts can seriously hamper downstream image processing tasks and hinder detection of subtle phenotypes. It is therefore of primary concern, and a prerequisite, to use quality control in HCS. In this work, we evaluate deep learning options that do not require extensive image annotations to provide a straightforward and easy to use semi-supervised learning solution to this issue. Concretely, we compared the efficacy of recent self-supervised and transfer learning approaches to provide a base encoder to a high throughput artefact image detector. The results of this study suggest that transfer learning methods should be preferred for this task as they not only performed best here but present the advantage of not requiring sensitive hyperparameter settings nor extensive additional training.
aura-net : robust segmentation of phase-contrast microscopy images with few annotations
Feb 02, 2021



Abstract:We present AURA-net, a convolutional neural network (CNN) for the segmentation of phase-contrast microscopy images. AURA-net uses transfer learning to accelerate training and Attention mechanisms to help the network focus on relevant image features. In this way, it can be trained efficiently with a very limited amount of annotations. Our network can thus be used to automate the segmentation of datasets that are generally considered too small for deep learning techniques. AURA-net also uses a loss inspired by active contours that is well-adapted to the specificity of phase-contrast images, further improving performance. We show that AURA-net outperforms state-of-the-art alternatives in several small (less than 100images) datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge