Nicolas Bourriez
XFACTORS: Disentangled Information Bottleneck via Contrastive Supervision
Jan 29, 2026Abstract:Disentangled representation learning aims to map independent factors of variation to independent representation components. On one hand, purely unsupervised approaches have proven successful on fully disentangled synthetic data, but fail to recover semantic factors from real data without strong inductive biases. On the other hand, supervised approaches are unstable and hard to scale to large attribute sets because they rely on adversarial objectives or auxiliary classifiers. We introduce \textsc{XFactors}, a weakly-supervised VAE framework that disentangles and provides explicit control over a chosen set of factors. Building on the Disentangled Information Bottleneck perspective, we decompose the representation into a residual subspace $\mathcal{S}$ and factor-specific subspaces $\mathcal{T}_1,\ldots,\mathcal{T}_K$ and a residual subspace $\mathcal{S}$. Each target factor is encoded in its assigned $\mathcal{T}_i$ through contrastive supervision: an InfoNCE loss pulls together latents sharing the same factor value and pushes apart mismatched pairs. In parallel, KL regularization imposes a Gaussian structure on both $\mathcal{S}$ and the aggregated factor subspaces, organizing the geometry without additional supervision for non-targeted factors and avoiding adversarial training and classifiers. Across multiple datasets, with constant hyperparameters, \textsc{XFactors} achieves state-of-the-art disentanglement scores and yields consistent qualitative factor alignment in the corresponding subspaces, enabling controlled factor swapping via latent replacement. We further demonstrate that our method scales correctly with increasing latent capacity and evaluate it on the real-world dataset CelebA. Our code is available at \href{https://github.com/ICML26-anon/XFactors}{github.com/ICML26-anon/XFactors}.
ChAda-ViT : Channel Adaptive Attention for Joint Representation Learning of Heterogeneous Microscopy Images
Nov 26, 2023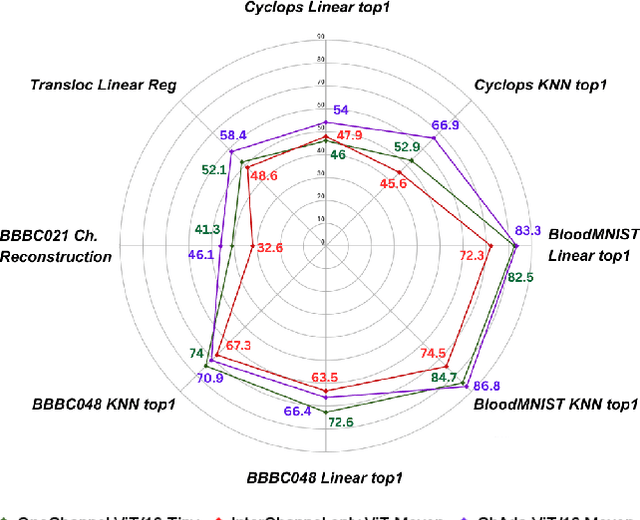
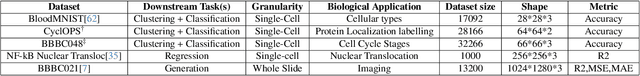
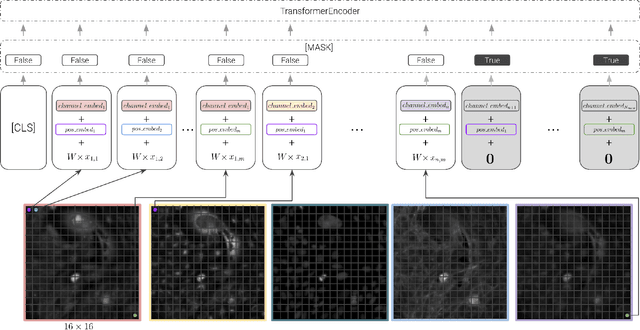

Abstract:Unlike color photography images, which are consistently encoded into RGB channels, biological images encompass various modalities, where the type of microscopy and the meaning of each channel varies with each experiment. Importantly, the number of channels can range from one to a dozen and their correlation is often comparatively much lower than RGB, as each of them brings specific information content. This aspect is largely overlooked by methods designed out of the bioimage field, and current solutions mostly focus on intra-channel spatial attention, often ignoring the relationship between channels, yet crucial in most biological applications. Importantly, the variable channel type and count prevent the projection of several experiments to a unified representation for large scale pre-training. In this study, we propose ChAda-ViT, a novel Channel Adaptive Vision Transformer architecture employing an Inter-Channel Attention mechanism on images with an arbitrary number, order and type of channels. We also introduce IDRCell100k, a bioimage dataset with a rich set of 79 experiments covering 7 microscope modalities, with a multitude of channel types, and channel counts varying from 1 to 10 per experiment. Our proposed architecture, trained in a self-supervised manner, outperforms existing approaches in several biologically relevant downstream tasks. Additionally, it can be used to bridge the gap for the first time between assays with different microscopes, channel numbers or types by embedding various image and experimental modalities into a unified biological image representation. The latter should facilitate interdisciplinary studies and pave the way for better adoption of deep learning in biological image-based analyses. Code and Data to be released soon.
Spacecraft Autonomous Decision-Planning for Collision Avoidance: a Reinforcement Learning Approach
Oct 29, 2023Abstract:The space environment around the Earth is becoming increasingly populated by both active spacecraft and space debris. To avoid potential collision events, significant improvements in Space Situational Awareness (SSA) activities and Collision Avoidance (CA) technologies are allowing the tracking and maneuvering of spacecraft with increasing accuracy and reliability. However, these procedures still largely involve a high level of human intervention to make the necessary decisions. For an increasingly complex space environment, this decision-making strategy is not likely to be sustainable. Therefore, it is important to successfully introduce higher levels of automation for key Space Traffic Management (STM) processes to ensure the level of reliability needed for navigating a large number of spacecraft. These processes range from collision risk detection to the identification of the appropriate action to take and the execution of avoidance maneuvers. This work proposes an implementation of autonomous CA decision-making capabilities on spacecraft based on Reinforcement Learning (RL) techniques. A novel methodology based on a Partially Observable Markov Decision Process (POMDP) framework is developed to train the Artificial Intelligence (AI) system on board the spacecraft, considering epistemic and aleatory uncertainties. The proposed framework considers imperfect monitoring information about the status of the debris in orbit and allows the AI system to effectively learn stochastic policies to perform accurate Collision Avoidance Maneuvers (CAMs). The objective is to successfully delegate the decision-making process for autonomously implementing a CAM to the spacecraft without human intervention. This approach would allow for a faster response in the decision-making process and for highly decentralized operations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge