Anthony Jennings
SOS: Selective Objective Switch for Rapid Immunofluorescence Whole Slide Image Classification
Mar 11, 2020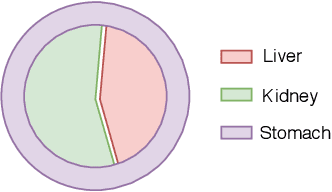
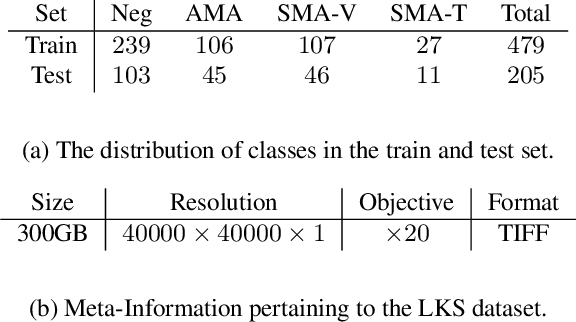
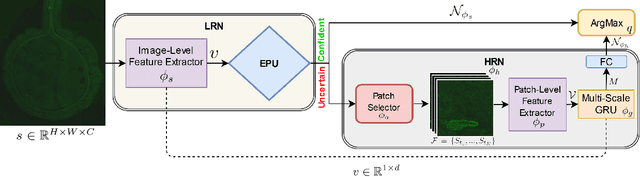
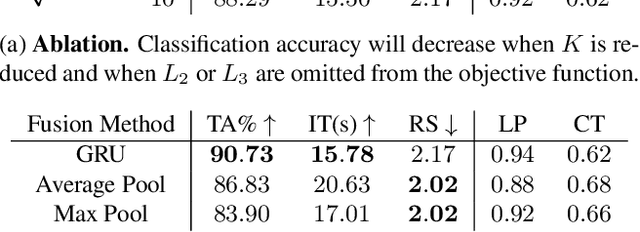
Abstract:The difficulty of processing gigapixel whole slide images (WSIs) in clinical microscopy has been a long-standing barrier to implementing computer aided diagnostic systems. Since modern computing resources are unable to perform computations at this extremely large scale, current state of the art methods utilize patch-based processing to preserve the resolution of WSIs. However, these methods are often resource intensive and make significant compromises on processing time. In this paper, we demonstrate that conventional patch-based processing is redundant for certain WSI classification tasks where high resolution is only required in a minority of cases. This reflects what is observed in clinical practice; where a pathologist may screen slides using a low power objective and only switch to a high power in cases where they are uncertain about their findings. To eliminate these redundancies, we propose a method for the selective use of high resolution processing based on the confidence of predictions on downscaled WSIs --- we call this the Selective Objective Switch (SOS). Our method is validated on a novel dataset of 684 Liver-Kidney-Stomach immunofluorescence WSIs routinely used in the investigation of autoimmune liver disease. By limiting high resolution processing to cases which cannot be classified confidently at low resolution, we maintain the accuracy of patch-level analysis whilst reducing the inference time by a factor of 7.74.
To What Extent Does Downsampling, Compression, and Data Scarcity Impact Renal Image Analysis?
Sep 22, 2019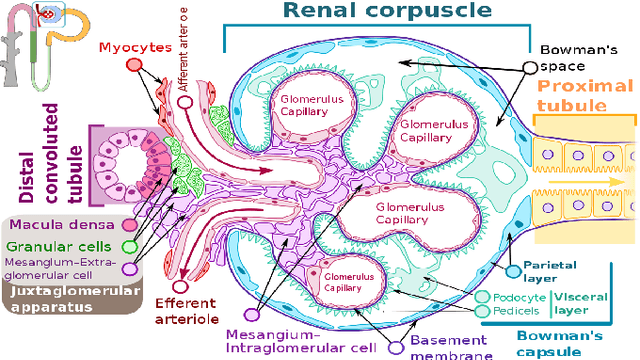
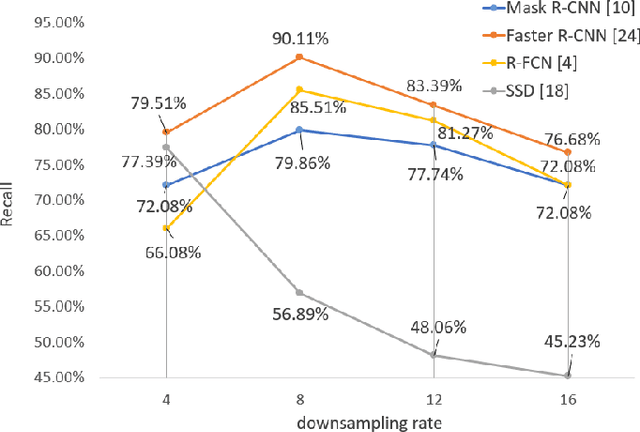
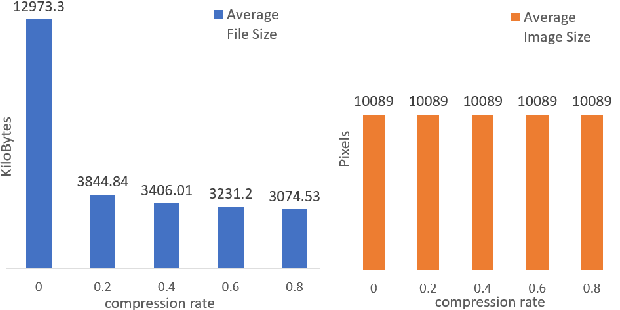
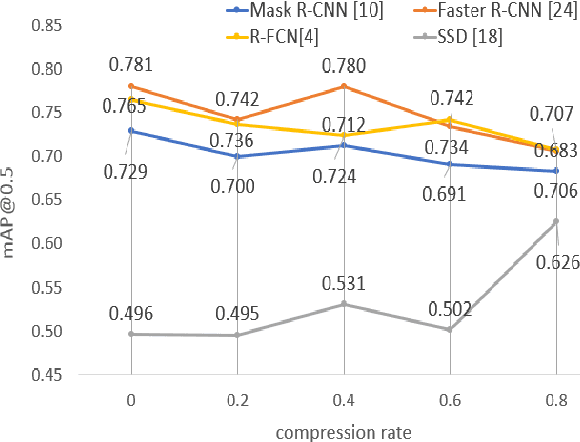
Abstract:The condition of the Glomeruli, or filter sacks, in renal Direct Immunofluorescence (DIF) specimens is a critical indicator for diagnosing kidney diseases. A digital pathology system which digitizes a glass histology slide into a Whole Slide Image (WSI) and then automatically detects and zooms in on the glomeruli with a higher magnification objective will be extremely helpful for pathologists. In this paper, using glomerulus detection as the study case, we provide analysis and observations on several important issues to help with the development of Computer Aided Diagnostic (CAD) systems to process WSIs. Large image resolution, large file size, and data scarcity are always challenging to deal with. To this end, we first examine image downsampling rates in terms of their effect on detection accuracy. Second, we examine the impact of image compression. Third, we examine the relationship between the size of the training set and detection accuracy. To understand the above issues, experiments are performed on the state-of-the-art detectors: Faster R-CNN, R-FCN, Mask R-CNN and SSD. Critical findings are observed: (1) The best balance between detection accuracy, detection speed and file size is achieved at 8 times downsampling captured with a $40\times$ objective; (2) compression which reduces the file size dramatically, does not necessarily have an adverse effect on overall accuracy; (3) reducing the amount of training data to some extents causes a drop in precision but has a negligible impact on the recall; (4) in most cases, Faster R-CNN achieves the best accuracy in the glomerulus detection task. We show that the image file size of $40\times$ WSI images can be reduced by a factor of over 6000 with negligible loss of glomerulus detection accuracy.
SlideNet: Fast and Accurate Slide Quality Assessment Based on Deep Neural Networks
Mar 20, 2018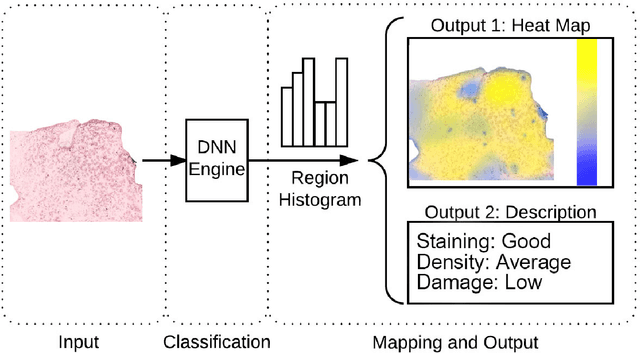

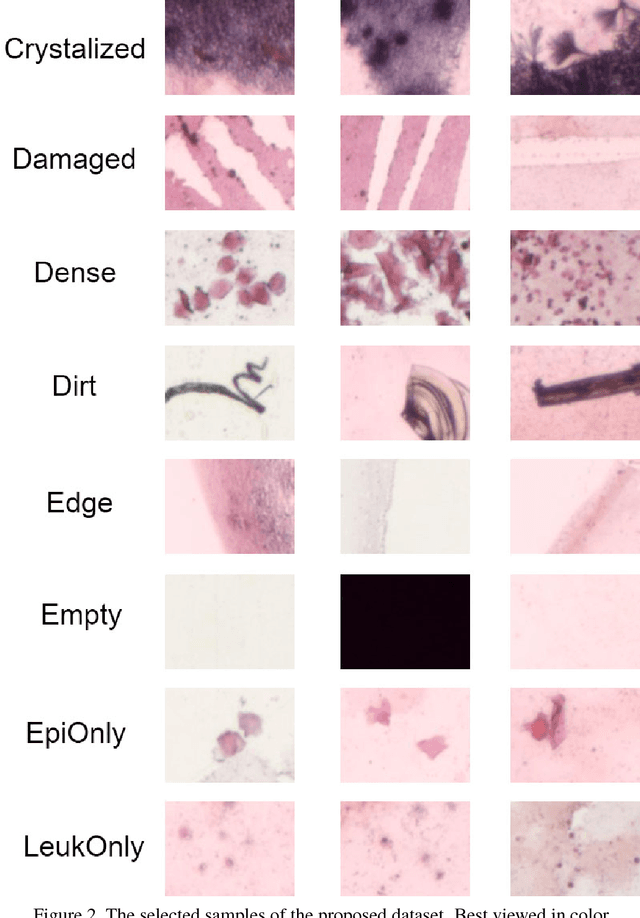
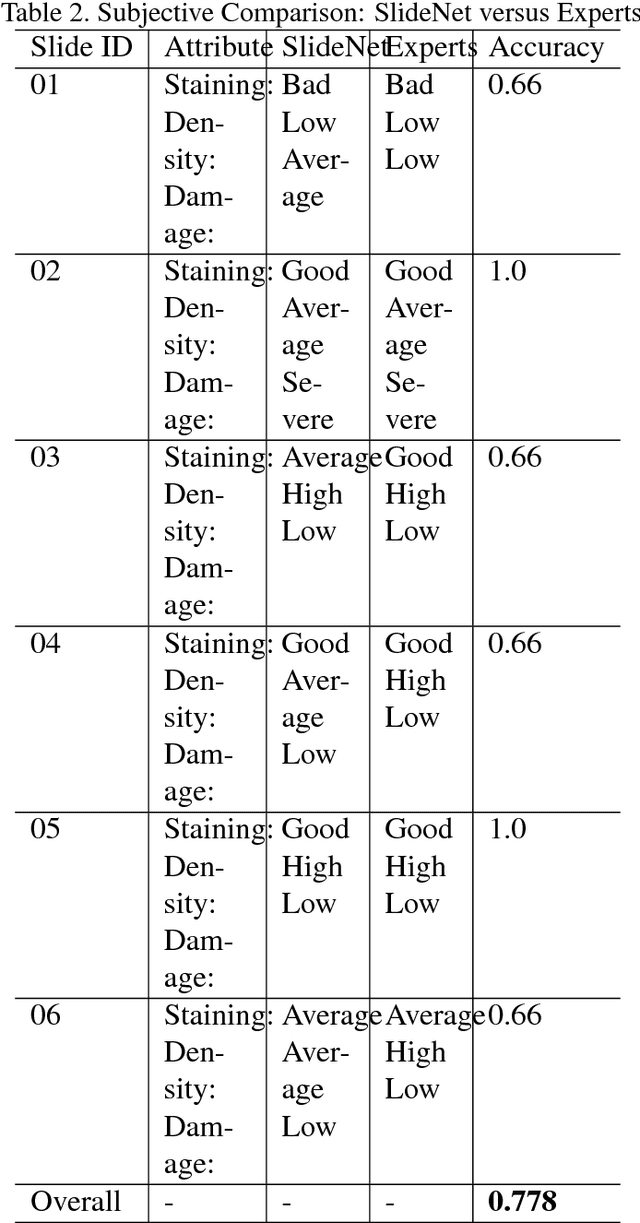
Abstract:This work tackles the automatic fine-grained slide quality assessment problem for digitized direct smears test using the Gram staining protocol. Automatic quality assessment can provide useful information for the pathologists and the whole digital pathology workflow. For instance, if the system found a slide to have a low staining quality, it could send a request to the automatic slide preparation system to remake the slide. If the system detects severe damage in the slides, it could notify the experts that manual microscope reading may be required. In order to address the quality assessment problem, we propose a deep neural network based framework to automatically assess the slide quality in a semantic way. Specifically, the first step of our framework is to perform dense fine-grained region classification on the whole slide and calculate the region distribution histogram. Next, our framework will generate assessments of the slide quality from various perspectives: staining quality, information density, damage level and which regions are more valuable for subsequent high-magnification analysis. To make the information more accessible, we present our results in the form of a heat map and text summaries. Additionally, in order to stimulate research in this direction, we propose a novel dataset for slide quality assessment. Experiments show that the proposed framework outperforms recent related works.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge