SOS: Selective Objective Switch for Rapid Immunofluorescence Whole Slide Image Classification
Paper and Code
Mar 11, 2020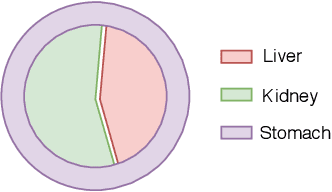
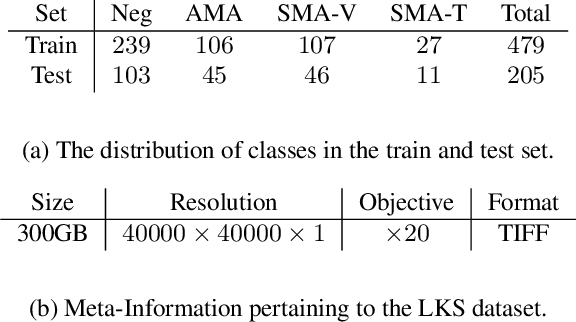
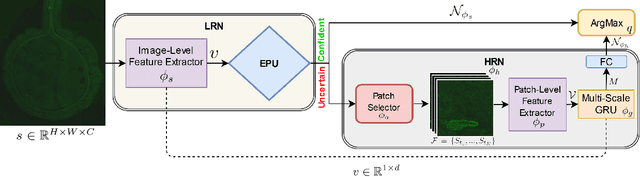
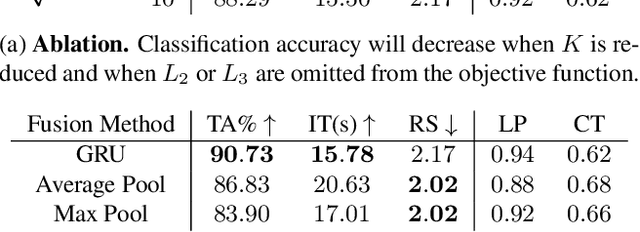
The difficulty of processing gigapixel whole slide images (WSIs) in clinical microscopy has been a long-standing barrier to implementing computer aided diagnostic systems. Since modern computing resources are unable to perform computations at this extremely large scale, current state of the art methods utilize patch-based processing to preserve the resolution of WSIs. However, these methods are often resource intensive and make significant compromises on processing time. In this paper, we demonstrate that conventional patch-based processing is redundant for certain WSI classification tasks where high resolution is only required in a minority of cases. This reflects what is observed in clinical practice; where a pathologist may screen slides using a low power objective and only switch to a high power in cases where they are uncertain about their findings. To eliminate these redundancies, we propose a method for the selective use of high resolution processing based on the confidence of predictions on downscaled WSIs --- we call this the Selective Objective Switch (SOS). Our method is validated on a novel dataset of 684 Liver-Kidney-Stomach immunofluorescence WSIs routinely used in the investigation of autoimmune liver disease. By limiting high resolution processing to cases which cannot be classified confidently at low resolution, we maintain the accuracy of patch-level analysis whilst reducing the inference time by a factor of 7.74.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge