Andrea Schenk
Robust Segmentation Models using an Uncertainty Slice Sampling Based Annotation Workflow
Sep 30, 2021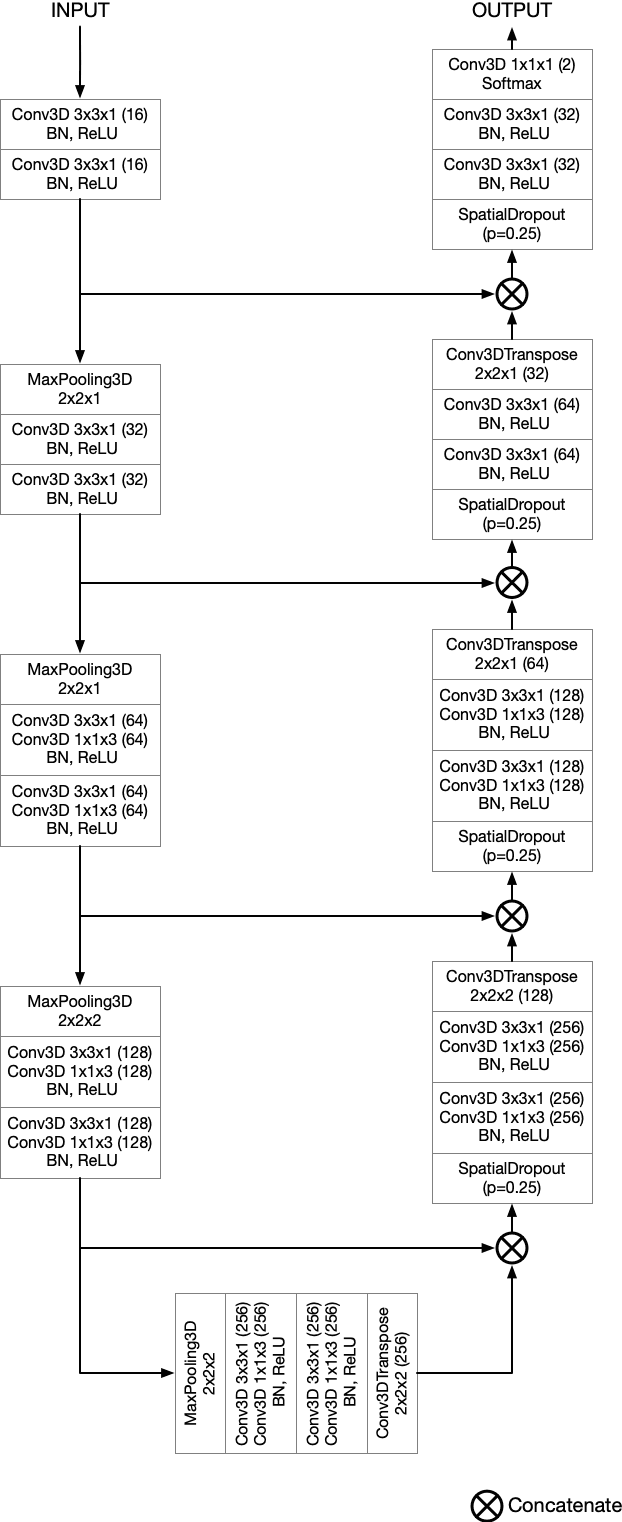
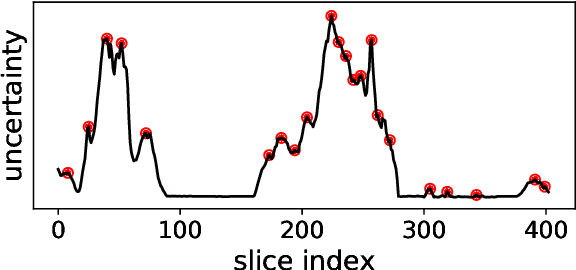
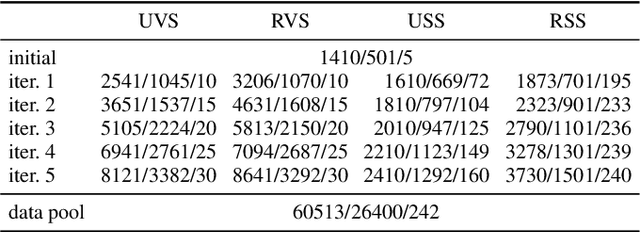
Abstract:Semantic segmentation neural networks require pixel-level annotations in large quantities to achieve a good performance. In the medical domain, such annotations are expensive, because they are time-consuming and require expert knowledge. Active learning optimizes the annotation effort by devising strategies to select cases for labeling that are most informative to the model. In this work, we propose an uncertainty slice sampling (USS) strategy for semantic segmentation of 3D medical volumes that selects 2D image slices for annotation and compare it with various other strategies. We demonstrate the efficiency of USS on a CT liver segmentation task using multi-site data. After five iterations, the training data resulting from USS consisted of 2410 slices (4% of all slices in the data pool) compared to 8121 (13%), 8641 (14%), and 3730 (6%) for uncertainty volume (UVS), random volume (RVS), and random slice (RSS) sampling, respectively. Despite being trained on the smallest amount of data, the model based on the USS strategy evaluated on 234 test volumes significantly outperformed models trained according to other strategies and achieved a mean Dice index of 0.964, a relative volume error of 4.2%, a mean surface distance of 1.35 mm, and a Hausdorff distance of 23.4 mm. This was only slightly inferior to 0.967, 3.8%, 1.18 mm, and 22.9 mm achieved by a model trained on all available data, but the robustness analysis using the 5th percentile of Dice and the 95th percentile of the remaining metrics demonstrated that USS resulted not only in the most robust model compared to other sampling schemes, but also outperformed the model trained on all data according to Dice (0.946 vs. 0.945) and mean surface distance (1.92 mm vs. 2.03 mm).
Anisotropic 3D Multi-Stream CNN for Accurate Prostate Segmentation from Multi-Planar MRI
Sep 23, 2020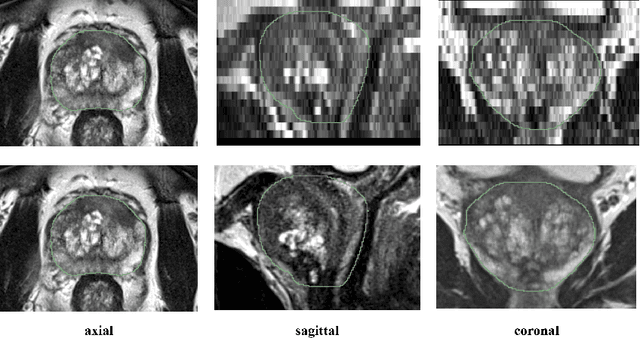
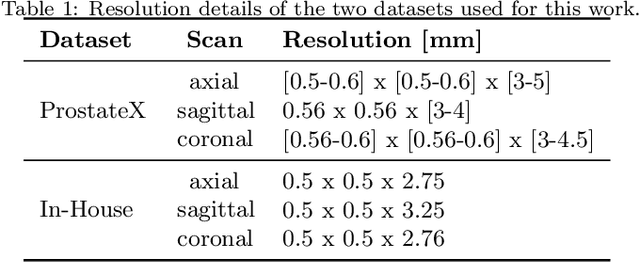
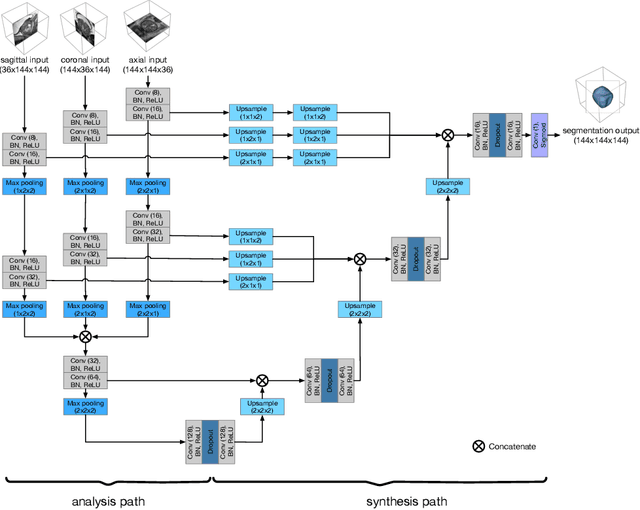
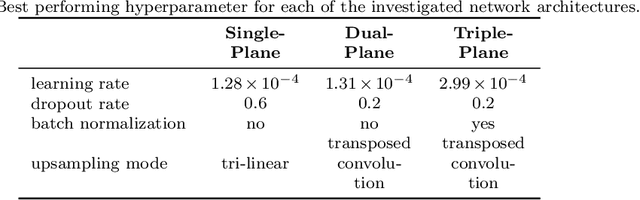
Abstract:Background and Objective: Accurate and reliable segmentation of the prostate gland in MR images can support the clinical assessment of prostate cancer, as well as the planning and monitoring of focal and loco-regional therapeutic interventions. Despite the availability of multi-planar MR scans due to standardized protocols, the majority of segmentation approaches presented in the literature consider the axial scans only. Methods: We propose an anisotropic 3D multi-stream CNN architecture, which processes additional scan directions to produce a higher-resolution isotropic prostate segmentation. We investigate two variants of our architecture, which work on two (dual-plane) and three (triple-plane) image orientations, respectively. We compare them with the standard baseline (single-plane) used in literature, i.e., plain axial segmentation. To realize a fair comparison, we employ a hyperparameter optimization strategy to select optimal configurations for the individual approaches. Results: Training and evaluation on two datasets spanning multiple sites obtain statistical significant improvement over the plain axial segmentation ($p<0.05$ on the Dice similarity coefficient). The improvement can be observed especially at the base ($0.898$ single-plane vs. $0.906$ triple-plane) and apex ($0.888$ single-plane vs. $0.901$ dual-plane). Conclusion: This study indicates that models employing two or three scan directions are superior to plain axial segmentation. The knowledge of precise boundaries of the prostate is crucial for the conservation of risk structures. Thus, the proposed models have the potential to improve the outcome of prostate cancer diagnosis and therapies.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019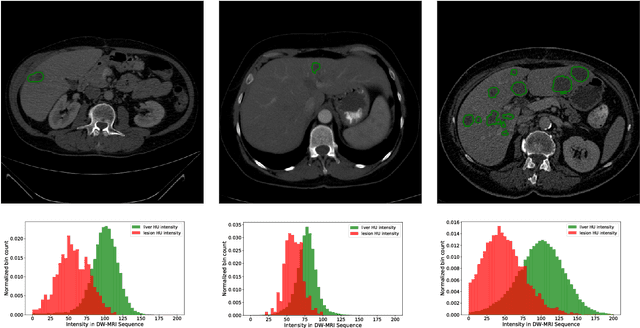
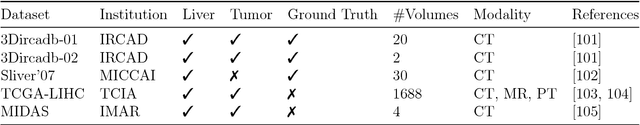
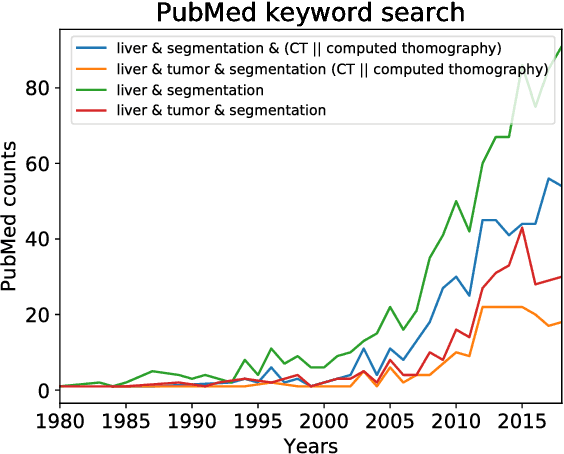
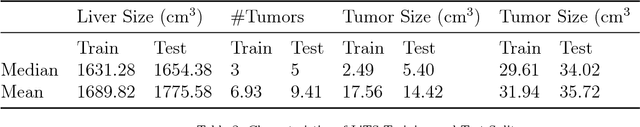
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Comparison of U-net-based Convolutional Neural Networks for Liver Segmentation in CT
Oct 09, 2018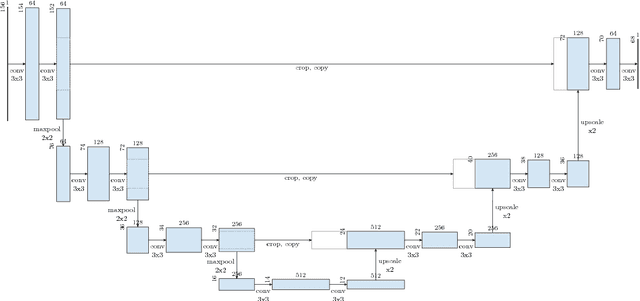
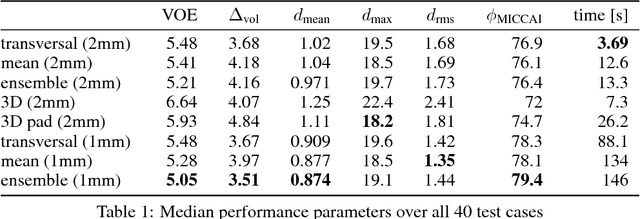
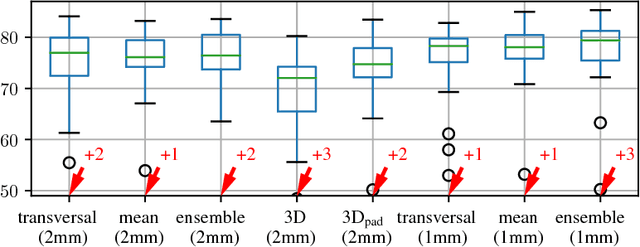

Abstract:Various approaches for liver segmentation in CT have been proposed: Besides statistical shape models, which played a major role in this research area, novel approaches on the basis of convolutional neural networks have been introduced recently. Using a set of 219 liver CT datasets with reference segmentations from liver surgery planning, we evaluate the performance of several neural network classifiers based on 2D and 3D U-net architectures. An interesting observation is that slice-wise approaches perform surprisingly well, with mean and median Dice coefficients above 0.97, and may be preferable over 3D approaches given current hardware and software limitations.
Neural Network-Based Automatic Liver Tumor Segmentation With Random Forest-Based Candidate Filtering
Jun 27, 2017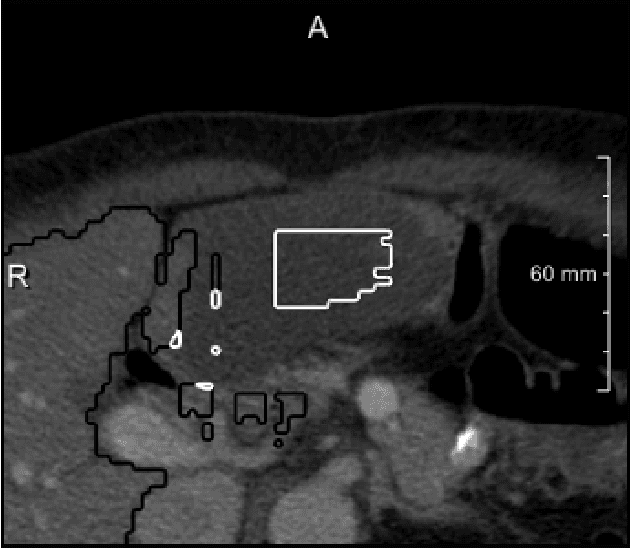
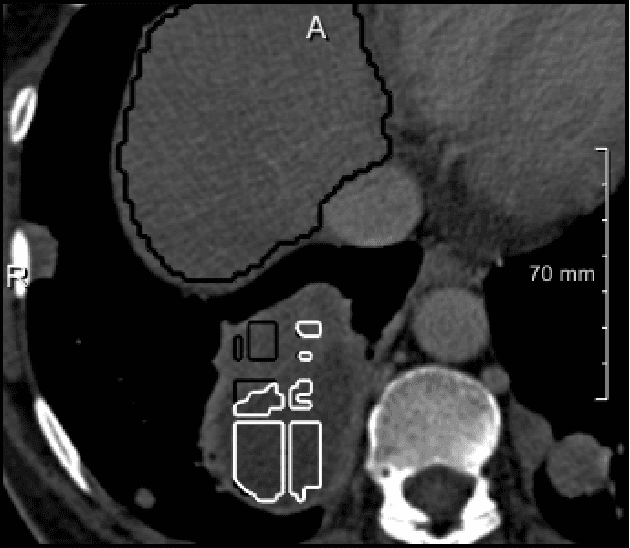
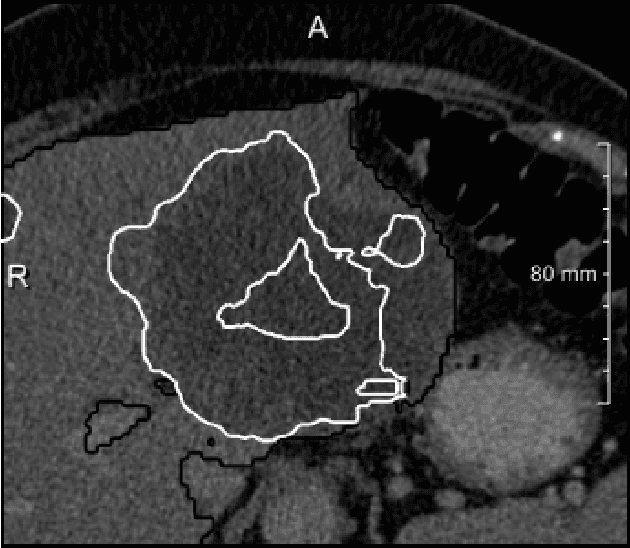
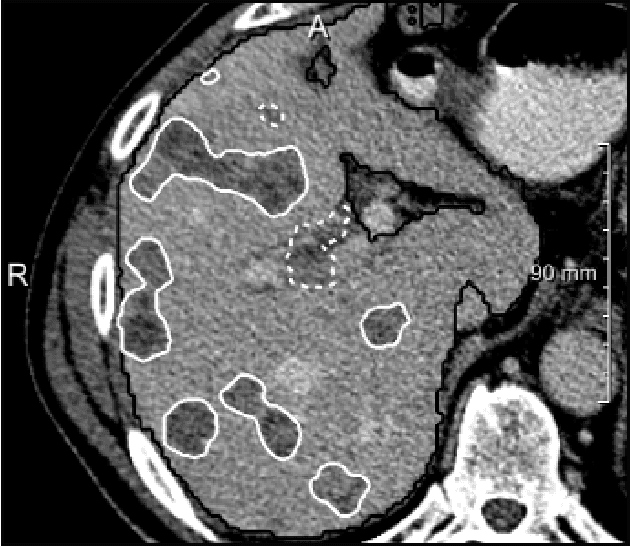
Abstract:We present a fully automatic method employing convolutional neural networks based on the 2D U-net architecture and random forest classifier to solve the automatic liver lesion segmentation problem of the ISBI 2017 Liver Tumor Segmentation Challenge (LiTS). In order to constrain the ROI in which the tumors could be located, a liver segmentation is performed first. For the organ segmentation, an ensemble of convolutional networks is trained to segment a liver using a set of 179 liver CT datasets from liver surgery planning. Inside of the liver ROI a neural network, trained using 127 challenge training datasets, identifies tumor candidates, which are subsequently filtered with a random forest classifier yielding the final tumor segmentation. The evaluation on the 70 challenge test cases resulted in a mean Dice coefficient of 0.65, ranking our method in the second place.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge