Robust Segmentation Models using an Uncertainty Slice Sampling Based Annotation Workflow
Paper and Code
Sep 30, 2021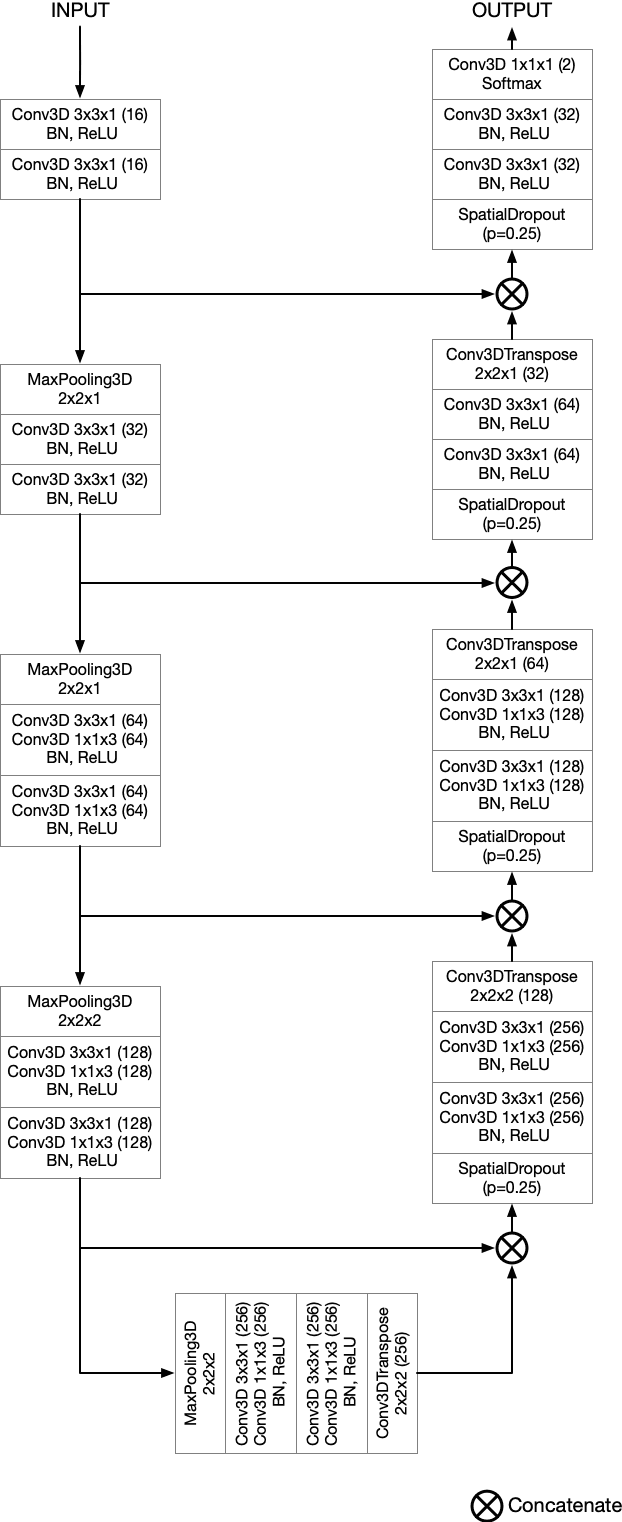
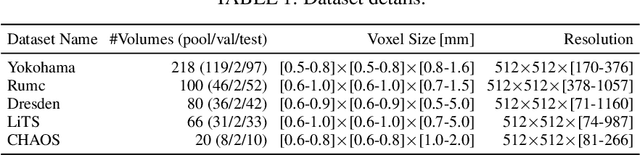
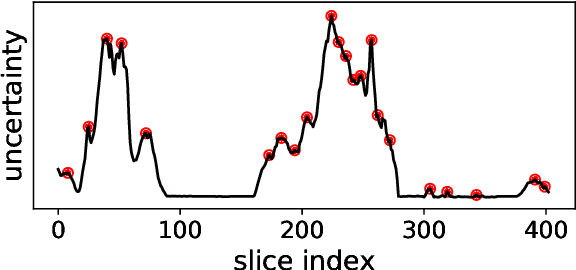
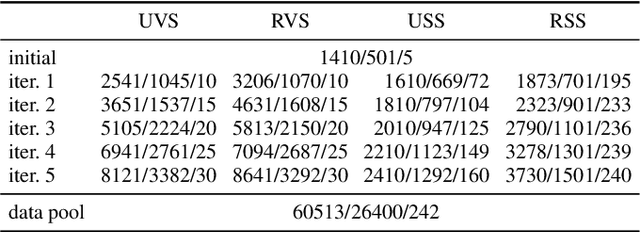
Semantic segmentation neural networks require pixel-level annotations in large quantities to achieve a good performance. In the medical domain, such annotations are expensive, because they are time-consuming and require expert knowledge. Active learning optimizes the annotation effort by devising strategies to select cases for labeling that are most informative to the model. In this work, we propose an uncertainty slice sampling (USS) strategy for semantic segmentation of 3D medical volumes that selects 2D image slices for annotation and compare it with various other strategies. We demonstrate the efficiency of USS on a CT liver segmentation task using multi-site data. After five iterations, the training data resulting from USS consisted of 2410 slices (4% of all slices in the data pool) compared to 8121 (13%), 8641 (14%), and 3730 (6%) for uncertainty volume (UVS), random volume (RVS), and random slice (RSS) sampling, respectively. Despite being trained on the smallest amount of data, the model based on the USS strategy evaluated on 234 test volumes significantly outperformed models trained according to other strategies and achieved a mean Dice index of 0.964, a relative volume error of 4.2%, a mean surface distance of 1.35 mm, and a Hausdorff distance of 23.4 mm. This was only slightly inferior to 0.967, 3.8%, 1.18 mm, and 22.9 mm achieved by a model trained on all available data, but the robustness analysis using the 5th percentile of Dice and the 95th percentile of the remaining metrics demonstrated that USS resulted not only in the most robust model compared to other sampling schemes, but also outperformed the model trained on all data according to Dice (0.946 vs. 0.945) and mean surface distance (1.92 mm vs. 2.03 mm).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge