Marko Rak
Predicting 4D Liver MRI for MR-guided Interventions
Feb 25, 2022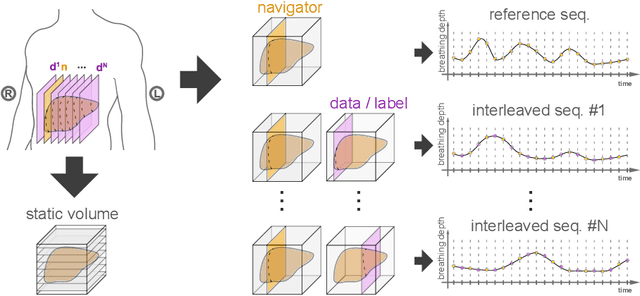

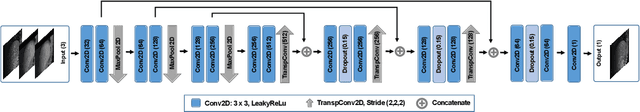
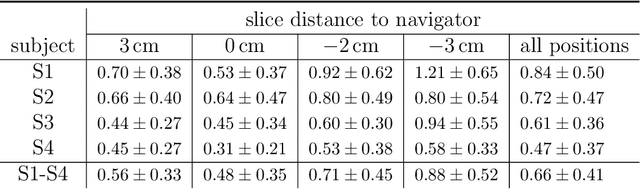
Abstract:Organ motion poses an unresolved challenge in image-guided interventions. In the pursuit of solving this problem, the research field of time-resolved volumetric magnetic resonance imaging (4D MRI) has evolved. However, current techniques are unsuitable for most interventional settings because they lack sufficient temporal and/or spatial resolution or have long acquisition times. In this work, we propose a novel approach for real-time, high-resolution 4D MRI with large fields of view for MR-guided interventions. To this end, we trained a convolutional neural network (CNN) end-to-end to predict a 3D liver MRI that correctly predicts the liver's respiratory state from a live 2D navigator MRI of a subject. Our method can be used in two ways: First, it can reconstruct near real-time 4D MRI with high quality and high resolution (209x128x128 matrix size with isotropic 1.8mm voxel size and 0.6s/volume) given a dynamic interventional 2D navigator slice for guidance during an intervention. Second, it can be used for retrospective 4D reconstruction with a temporal resolution of below 0.2s/volume for motion analysis and use in radiation therapy. We report a mean target registration error (TRE) of 1.19 $\pm$0.74mm, which is below voxel size. We compare our results with a state-of-the-art retrospective 4D MRI reconstruction. Visual evaluation shows comparable quality. We show that small training sizes with short acquisition times down to 2min can already achieve promising results and 24min are sufficient for high quality results. Because our method can be readily combined with earlier methods, acquisition time can be further decreased while also limiting quality loss. We show that an end-to-end, deep learning formulation is highly promising for 4D MRI reconstruction.
Uncertainty-Aware Temporal Self-Learning (UATS): Semi-Supervised Learning for Segmentation of Prostate Zones and Beyond
Apr 08, 2021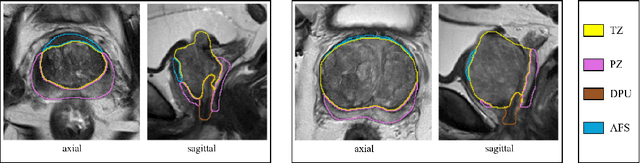
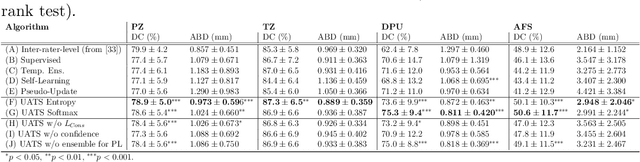
Abstract:Various convolutional neural network (CNN) based concepts have been introduced for the prostate's automatic segmentation and its coarse subdivision into transition zone (TZ) and peripheral zone (PZ). However, when targeting a fine-grained segmentation of TZ, PZ, distal prostatic urethra (DPU) and the anterior fibromuscular stroma (AFS), the task becomes more challenging and has not yet been solved at the level of human performance. One reason might be the insufficient amount of labeled data for supervised training. Therefore, we propose to apply a semi-supervised learning (SSL) technique named uncertainty-aware temporal self-learning (UATS) to overcome the expensive and time-consuming manual ground truth labeling. We combine the SSL techniques temporal ensembling and uncertainty-guided self-learning to benefit from unlabeled images, which are often readily available. Our method significantly outperforms the supervised baseline and obtained a Dice coefficient (DC) of up to 78.9% , 87.3%, 75.3%, 50.6% for TZ, PZ, DPU and AFS, respectively. The obtained results are in the range of human inter-rater performance for all structures. Moreover, we investigate the method's robustness against noise and demonstrate the generalization capability for varying ratios of labeled data and on other challenging tasks, namely the hippocampus and skin lesion segmentation. UATS achieved superiority segmentation quality compared to the supervised baseline, particularly for minimal amounts of labeled data.
Learning Multi-Modal Volumetric Prostate Registration with Weak Inter-Subject Spatial Correspondence
Feb 09, 2021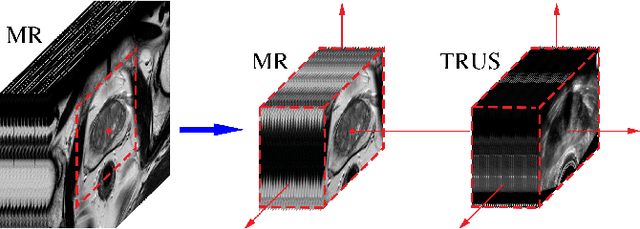
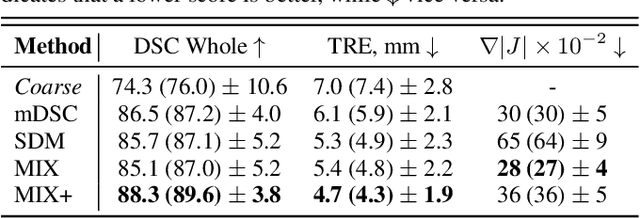
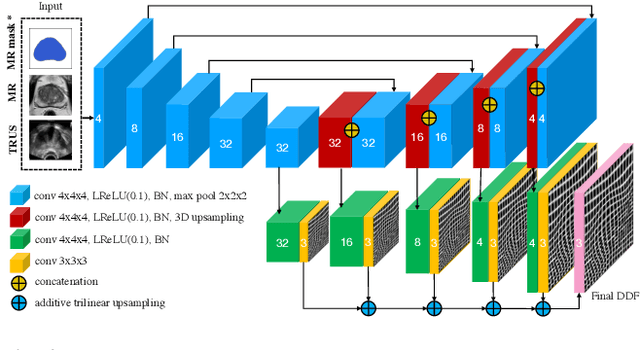
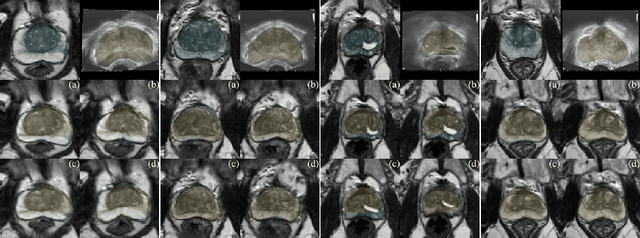
Abstract:Recent studies demonstrated the eligibility of convolutional neural networks (CNNs) for solving the image registration problem. CNNs enable faster transformation estimation and greater generalization capability needed for better support during medical interventions. Conventional fully-supervised training requires a lot of high-quality ground truth data such as voxel-to-voxel transformations, which typically are attained in a too tedious and error-prone manner. In our work, we use weakly-supervised learning, which optimizes the model indirectly only via segmentation masks that are a more accessible ground truth than the deformation fields. Concerning the weak supervision, we investigate two segmentation similarity measures: multiscale Dice similarity coefficient (mDSC) and the similarity between segmentation-derived signed distance maps (SDMs). We show that the combination of mDSC and SDM similarity measures results in a more accurate and natural transformation pattern together with a stronger gradient coverage. Furthermore, we introduce an auxiliary input to the neural network for the prior information about the prostate location in the MR sequence, which mostly is available preoperatively. This approach significantly outperforms the standard two-input models. With weakly labelled MR-TRUS prostate data, we showed registration quality comparable to the state-of-the-art deep learning-based method.
Anisotropic 3D Multi-Stream CNN for Accurate Prostate Segmentation from Multi-Planar MRI
Sep 23, 2020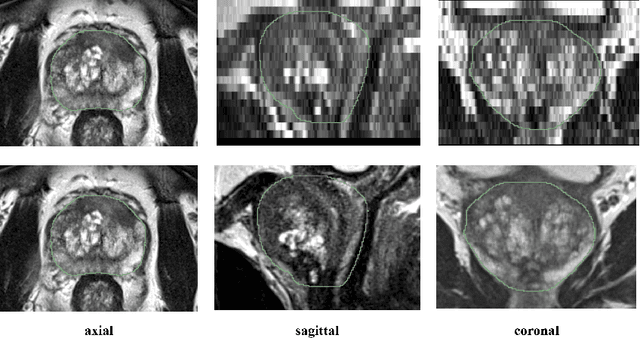
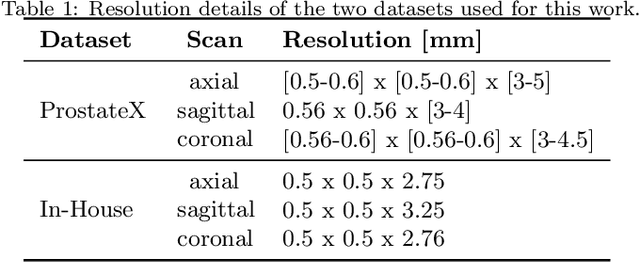
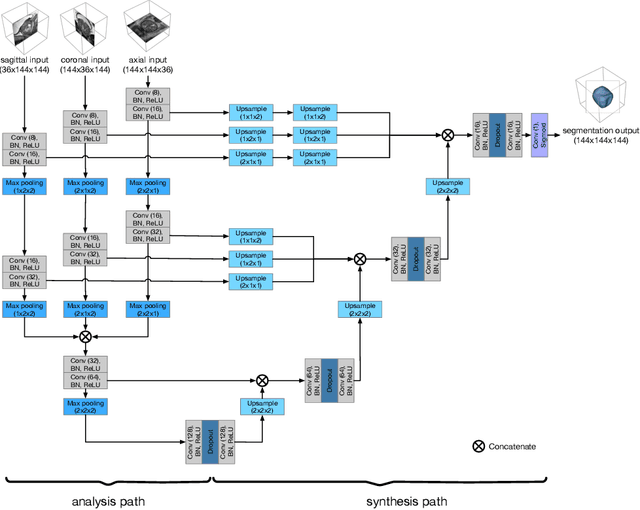
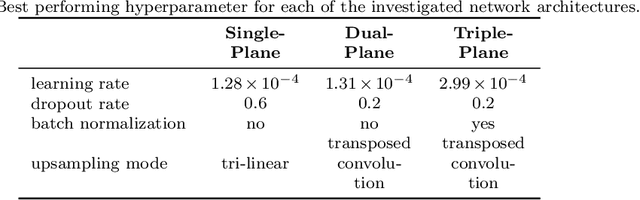
Abstract:Background and Objective: Accurate and reliable segmentation of the prostate gland in MR images can support the clinical assessment of prostate cancer, as well as the planning and monitoring of focal and loco-regional therapeutic interventions. Despite the availability of multi-planar MR scans due to standardized protocols, the majority of segmentation approaches presented in the literature consider the axial scans only. Methods: We propose an anisotropic 3D multi-stream CNN architecture, which processes additional scan directions to produce a higher-resolution isotropic prostate segmentation. We investigate two variants of our architecture, which work on two (dual-plane) and three (triple-plane) image orientations, respectively. We compare them with the standard baseline (single-plane) used in literature, i.e., plain axial segmentation. To realize a fair comparison, we employ a hyperparameter optimization strategy to select optimal configurations for the individual approaches. Results: Training and evaluation on two datasets spanning multiple sites obtain statistical significant improvement over the plain axial segmentation ($p<0.05$ on the Dice similarity coefficient). The improvement can be observed especially at the base ($0.898$ single-plane vs. $0.906$ triple-plane) and apex ($0.888$ single-plane vs. $0.901$ dual-plane). Conclusion: This study indicates that models employing two or three scan directions are superior to plain axial segmentation. The knowledge of precise boundaries of the prostate is crucial for the conservation of risk structures. Thus, the proposed models have the potential to improve the outcome of prostate cancer diagnosis and therapies.
4D MRI: Robust sorting of free breathing MRI slices for use in interventional settings
Oct 04, 2019
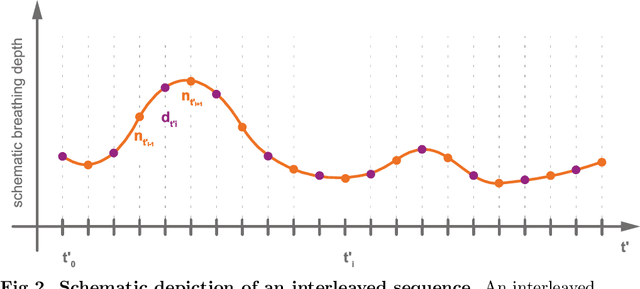
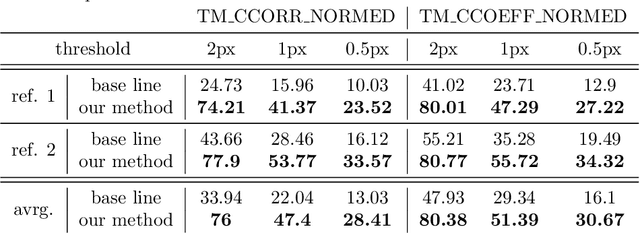
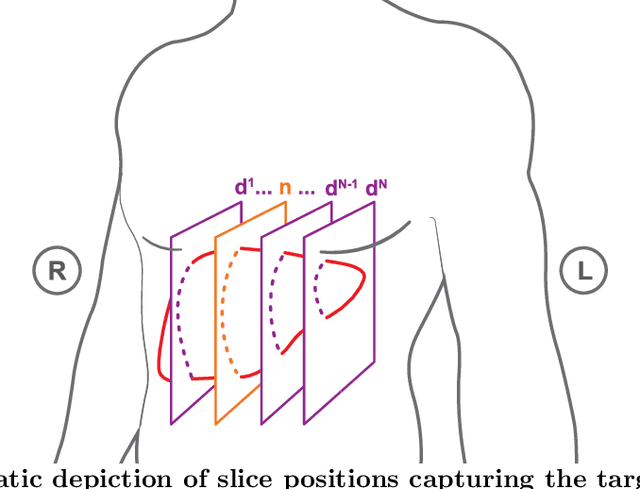
Abstract:Purpose: We aim to develop a robust 4D MRI method for large FOVs enabling the extraction of irregular respiratory motion that is readily usable with all MRI machines and thus applicable to support a wide range of interventional settings. Method: We propose a 4D MRI reconstruction method to capture an arbitrary number of breathing states. It uses template updates in navigator slices and search regions for fast and robust vessel cross-section tracking. It captures FOVs of 255 mm x 320 mm x 228 mm at a spatial resolution of 1.82 mm x 1.82 mm x 4mm and temporal resolution of 200ms. To validate the method, a total of 38 4D MRIs of 13 healthy subjects were reconstructed. A quantitative evaluation of the reconstruction rate and speed of both the new and baseline method was performed. Additionally, a study with ten radiologists was conducted to assess the subjective reconstruction quality of both methods. Results: Our results indicate improved mean reconstruction rates compared to the baseline method (79.4\% vs. 45.5\%) and improved mean reconstruction times (24s vs. 73s) per subject. Interventional radiologists perceive the reconstruction quality of our method as higher compared to the baseline (262.5 points vs. 217.5 points, p=0.02). Conclusions: Template updates are an effective and efficient way to increase 4D MRI reconstruction rates and to achieve better reconstruction quality. Search regions reduce reconstruction time. These improvements increase the applicability of 4D MRI as base for seamless support of interventional image guidance in percutaneous interventions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge