Oleksii Bashkanov
Predicting 4D Liver MRI for MR-guided Interventions
Feb 25, 2022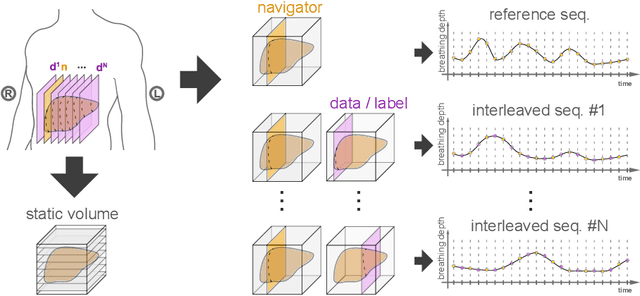

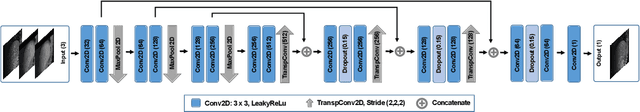
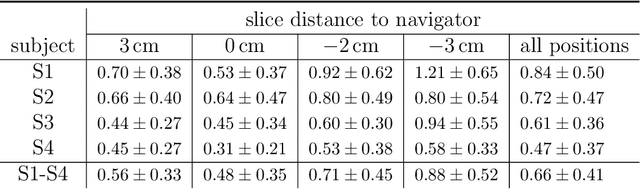
Abstract:Organ motion poses an unresolved challenge in image-guided interventions. In the pursuit of solving this problem, the research field of time-resolved volumetric magnetic resonance imaging (4D MRI) has evolved. However, current techniques are unsuitable for most interventional settings because they lack sufficient temporal and/or spatial resolution or have long acquisition times. In this work, we propose a novel approach for real-time, high-resolution 4D MRI with large fields of view for MR-guided interventions. To this end, we trained a convolutional neural network (CNN) end-to-end to predict a 3D liver MRI that correctly predicts the liver's respiratory state from a live 2D navigator MRI of a subject. Our method can be used in two ways: First, it can reconstruct near real-time 4D MRI with high quality and high resolution (209x128x128 matrix size with isotropic 1.8mm voxel size and 0.6s/volume) given a dynamic interventional 2D navigator slice for guidance during an intervention. Second, it can be used for retrospective 4D reconstruction with a temporal resolution of below 0.2s/volume for motion analysis and use in radiation therapy. We report a mean target registration error (TRE) of 1.19 $\pm$0.74mm, which is below voxel size. We compare our results with a state-of-the-art retrospective 4D MRI reconstruction. Visual evaluation shows comparable quality. We show that small training sizes with short acquisition times down to 2min can already achieve promising results and 24min are sufficient for high quality results. Because our method can be readily combined with earlier methods, acquisition time can be further decreased while also limiting quality loss. We show that an end-to-end, deep learning formulation is highly promising for 4D MRI reconstruction.
Learning Multi-Modal Volumetric Prostate Registration with Weak Inter-Subject Spatial Correspondence
Feb 09, 2021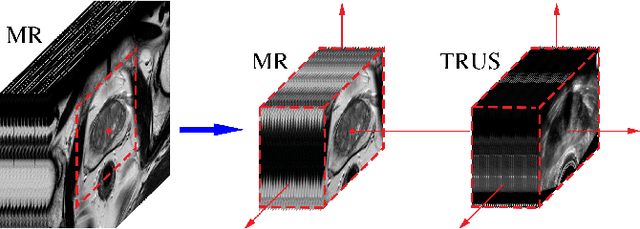
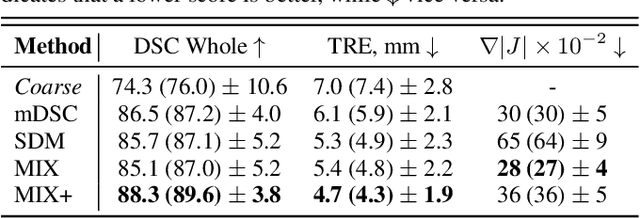
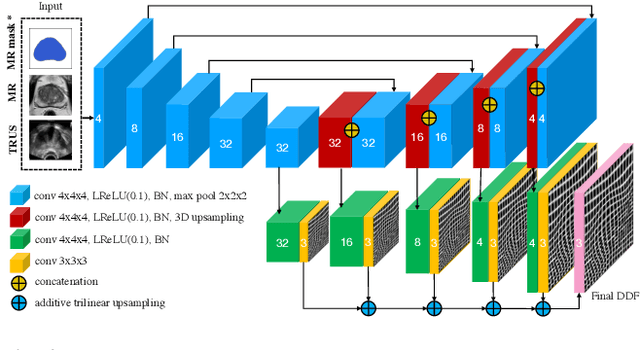
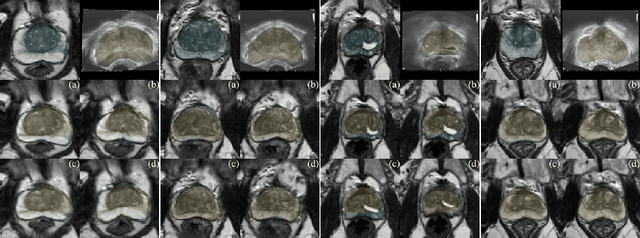
Abstract:Recent studies demonstrated the eligibility of convolutional neural networks (CNNs) for solving the image registration problem. CNNs enable faster transformation estimation and greater generalization capability needed for better support during medical interventions. Conventional fully-supervised training requires a lot of high-quality ground truth data such as voxel-to-voxel transformations, which typically are attained in a too tedious and error-prone manner. In our work, we use weakly-supervised learning, which optimizes the model indirectly only via segmentation masks that are a more accessible ground truth than the deformation fields. Concerning the weak supervision, we investigate two segmentation similarity measures: multiscale Dice similarity coefficient (mDSC) and the similarity between segmentation-derived signed distance maps (SDMs). We show that the combination of mDSC and SDM similarity measures results in a more accurate and natural transformation pattern together with a stronger gradient coverage. Furthermore, we introduce an auxiliary input to the neural network for the prior information about the prostate location in the MR sequence, which mostly is available preoperatively. This approach significantly outperforms the standard two-input models. With weakly labelled MR-TRUS prostate data, we showed registration quality comparable to the state-of-the-art deep learning-based method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge