Anders S. Christensen
NeuralPLexer3: Accurate Biomolecular Complex Structure Prediction with Flow Models
Dec 18, 2024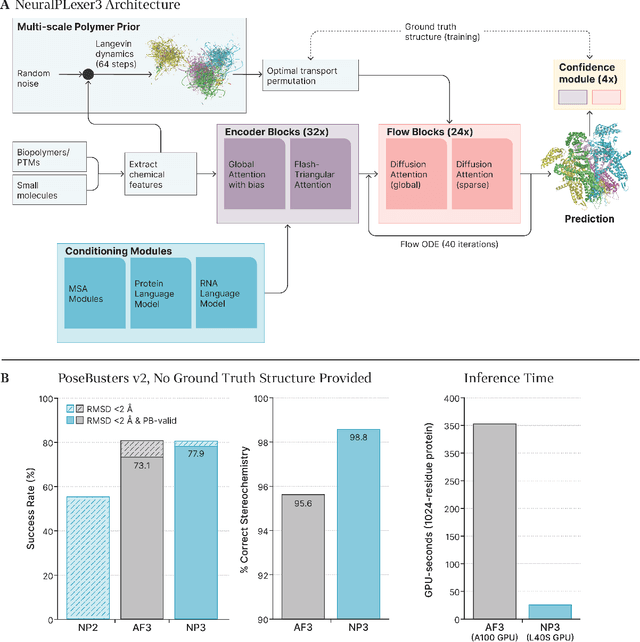
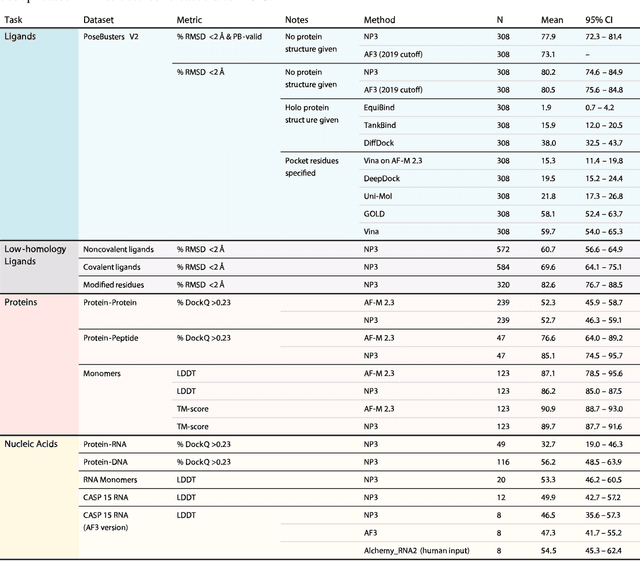
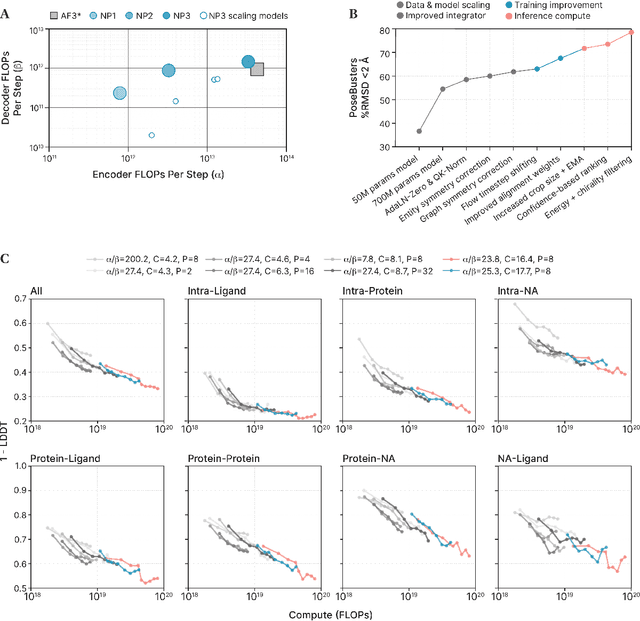
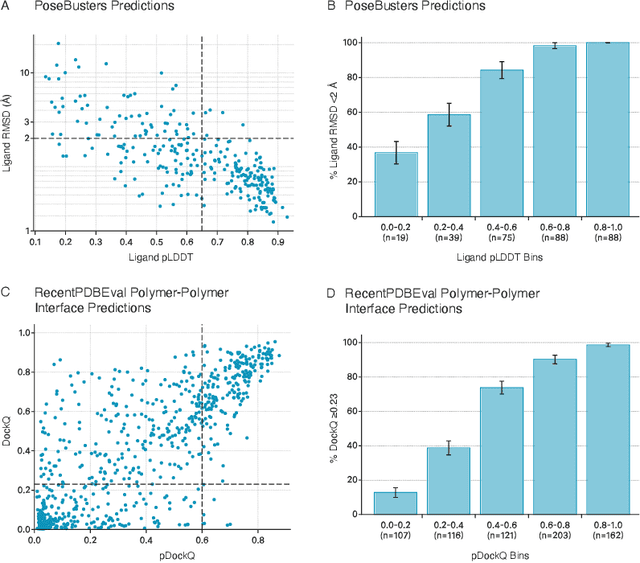
Abstract:Structure determination is essential to a mechanistic understanding of diseases and the development of novel therapeutics. Machine-learning-based structure prediction methods have made significant advancements by computationally predicting protein and bioassembly structures from sequences and molecular topology alone. Despite substantial progress in the field, challenges remain to deliver structure prediction models to real-world drug discovery. Here, we present NeuralPLexer3 -- a physics-inspired flow-based generative model that achieves state-of-the-art prediction accuracy on key biomolecular interaction types and improves training and sampling efficiency compared to its predecessors and alternative methodologies. Examined through newly developed benchmarking strategies, NeuralPLexer3 excels in vital areas that are crucial to structure-based drug design, such as physical validity and ligand-induced conformational changes.
UNiTE: Unitary N-body Tensor Equivariant Network with Applications to Quantum Chemistry
Jun 06, 2021
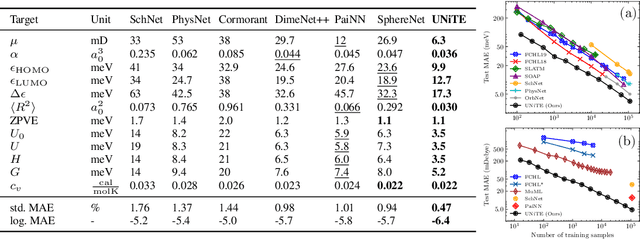
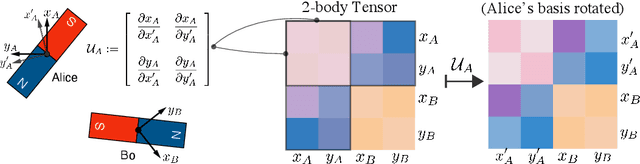
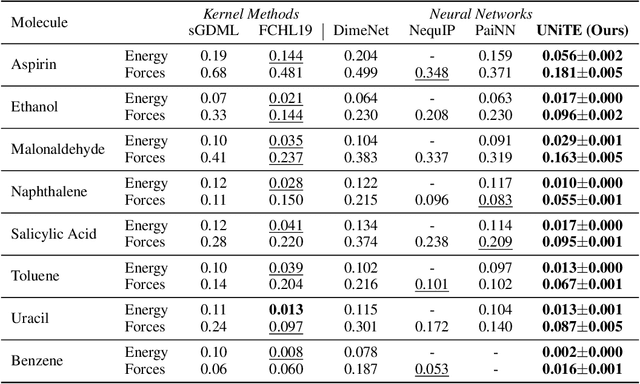
Abstract:Equivariant neural networks have been successful in incorporating various types of symmetries, but are mostly limited to vector representations of geometric objects. Despite the prevalence of higher-order tensors in various application domains, e.g. in quantum chemistry, equivariant neural networks for general tensors remain unexplored. Previous strategies for learning equivariant functions on tensors mostly rely on expensive tensor factorization which is not scalable when the dimensionality of the problem becomes large. In this work, we propose unitary $N$-body tensor equivariant neural network (UNiTE), an architecture for a general class of symmetric tensors called $N$-body tensors. The proposed neural network is equivariant with respect to the actions of a unitary group, such as the group of 3D rotations. Furthermore, it has a linear time complexity with respect to the number of non-zero elements in the tensor. We also introduce a normalization method, viz., Equivariant Normalization, to improve generalization of the neural network while preserving symmetry. When applied to quantum chemistry, UNiTE outperforms all state-of-the-art machine learning methods of that domain with over 110% average improvements on multiple benchmarks. Finally, we show that UNiTE achieves a robust zero-shot generalization performance on diverse down stream chemistry tasks, while being three orders of magnitude faster than conventional numerical methods with competitive accuracy.
Neural networks and kernel ridge regression for excited states dynamics of CH$_2$NH$_2^+$: From single-state to multi-state representations and multi-property machine learning models
Dec 18, 2019

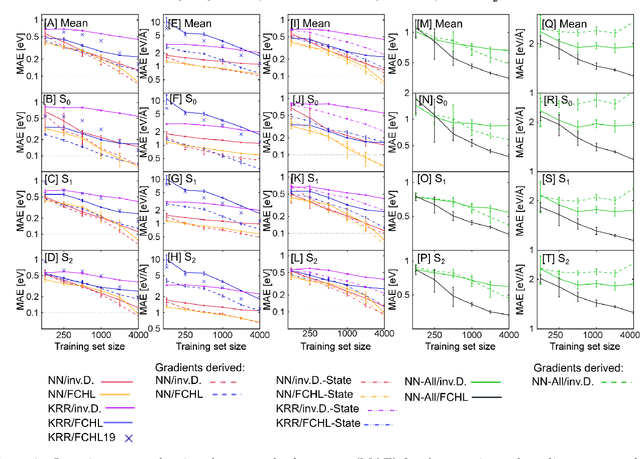

Abstract:Excited-state dynamics simulations are a powerful tool to investigate photo-induced reactions of molecules and materials and provide complementary information to experiments. Since the applicability of these simulation techniques is limited by the costs of the underlying electronic structure calculations, we develop and assess different machine learning models for this task. The machine learning models are trained on {\emph ab initio} calculations for excited electronic states, using the methylenimmonium cation (CH$_2$NH$_2^+$) as a model system. For the prediction of excited-state properties, multiple outputs are desirable, which is straightforward with neural networks but less explored with kernel ridge regression. We overcome this challenge for kernel ridge regression in the case of energy predictions by encoding the electronic states explicitly in the inputs, in addition to the molecular representation. We adopt this strategy also for our neural networks for comparison. Such a state encoding enables not only kernel ridge regression with multiple outputs but leads also to more accurate machine learning models for state-specific properties. An important goal for excited-state machine learning models is their use in dynamics simulations, which needs not only state-specific information but also couplings, i.e., properties involving pairs of states. Accordingly, we investigate the performance of different models for such coupling elements. Furthermore, we explore how combining all properties in a single neural network affects the accuracy. As an ultimate test for our machine learning models, we carry out excited-state dynamics simulations based on the predicted energies, forces and couplings and, thus, show the scopes and possibilities of machine learning for the treatment of electronically excited states.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge