Aleksandr Matsun
CoReEcho: Continuous Representation Learning for 2D+time Echocardiography Analysis
Mar 15, 2024



Abstract:Deep learning (DL) models have been advancing automatic medical image analysis on various modalities, including echocardiography, by offering a comprehensive end-to-end training pipeline. This approach enables DL models to regress ejection fraction (EF) directly from 2D+time echocardiograms, resulting in superior performance. However, the end-to-end training pipeline makes the learned representations less explainable. The representations may also fail to capture the continuous relation among echocardiogram clips, indicating the existence of spurious correlations, which can negatively affect the generalization. To mitigate this issue, we propose CoReEcho, a novel training framework emphasizing continuous representations tailored for direct EF regression. Our extensive experiments demonstrate that CoReEcho: 1) outperforms the current state-of-the-art (SOTA) on the largest echocardiography dataset (EchoNet-Dynamic) with MAE of 3.90 & R2 of 82.44, and 2) provides robust and generalizable features that transfer more effectively in related downstream tasks. The code is publicly available at https://github.com/fadamsyah/CoReEcho.
ConDiSR: Contrastive Disentanglement and Style Regularization for Single Domain Generalization
Mar 14, 2024Abstract:Medical data often exhibits distribution shifts, which cause test-time performance degradation for deep learning models trained using standard supervised learning pipelines. This challenge is addressed in the field of Domain Generalization (DG) with the sub-field of Single Domain Generalization (SDG) being specifically interesting due to the privacy- or logistics-related issues often associated with medical data. Existing disentanglement-based SDG methods heavily rely on structural information embedded in segmentation masks, however classification labels do not provide such dense information. This work introduces a novel SDG method aimed at medical image classification that leverages channel-wise contrastive disentanglement. It is further enhanced with reconstruction-based style regularization to ensure extraction of distinct style and structure feature representations. We evaluate our method on the complex task of multicenter histopathology image classification, comparing it against state-of-the-art (SOTA) SDG baselines. Results demonstrate that our method surpasses the SOTA by a margin of 1% in average accuracy while also showing more stable performance. This study highlights the importance and challenges of exploring SDG frameworks in the context of the classification task. The code is publicly available at https://github.com/BioMedIA-MBZUAI/ConDiSR
Innovative Horizons in Aerial Imagery: LSKNet Meets DiffusionDet for Advanced Object Detection
Nov 21, 2023Abstract:In the realm of aerial image analysis, object detection plays a pivotal role, with significant implications for areas such as remote sensing, urban planning, and disaster management. This study addresses the inherent challenges in this domain, notably the detection of small objects, managing densely packed elements, and accounting for diverse orientations. We present an in-depth evaluation of an object detection model that integrates the Large Selective Kernel Network (LSKNet)as its backbone with the DiffusionDet head, utilizing the iSAID dataset for empirical analysis. Our approach encompasses the introduction of novel methodologies and extensive ablation studies. These studies critically assess various aspects such as loss functions, box regression techniques, and classification strategies to refine the model's precision in object detection. The paper details the experimental application of the LSKNet backbone in synergy with the DiffusionDet heads, a combination tailored to meet the specific challenges in aerial image object detection. The findings of this research indicate a substantial enhancement in the model's performance, especially in the accuracy-time tradeoff. The proposed model achieves a mean average precision (MAP) of approximately 45.7%, which is a significant improvement, outperforming the RCNN model by 4.7% on the same dataset. This advancement underscores the effectiveness of the proposed modifications and sets a new benchmark in aerial image analysis, paving the way for more accurate and efficient object detection methodologies. The code is publicly available at https://github.com/SashaMatsun/LSKDiffDet
DGM-DR: Domain Generalization with Mutual Information Regularized Diabetic Retinopathy Classification
Sep 18, 2023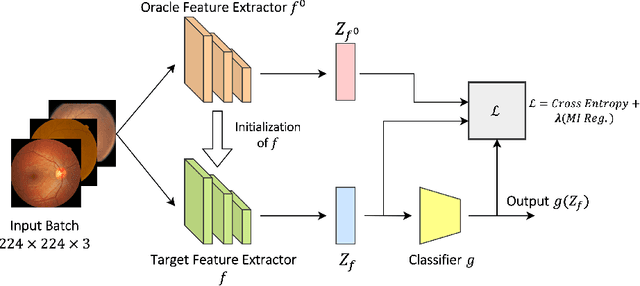
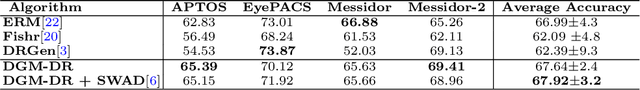
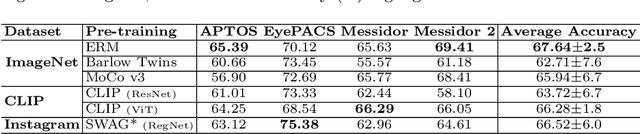
Abstract:The domain shift between training and testing data presents a significant challenge for training generalizable deep learning models. As a consequence, the performance of models trained with the independent and identically distributed (i.i.d) assumption deteriorates when deployed in the real world. This problem is exacerbated in the medical imaging context due to variations in data acquisition across clinical centers, medical apparatus, and patients. Domain generalization (DG) aims to address this problem by learning a model that generalizes well to any unseen target domain. Many domain generalization techniques were unsuccessful in learning domain-invariant representations due to the large domain shift. Furthermore, multiple tasks in medical imaging are not yet extensively studied in existing literature when it comes to DG point of view. In this paper, we introduce a DG method that re-establishes the model objective function as a maximization of mutual information with a large pretrained model to the medical imaging field. We re-visit the problem of DG in Diabetic Retinopathy (DR) classification to establish a clear benchmark with a correct model selection strategy and to achieve robust domain-invariant representation for an improved generalization. Moreover, we conduct extensive experiments on public datasets to show that our proposed method consistently outperforms the previous state-of-the-art by a margin of 5.25% in average accuracy and a lower standard deviation. Source code available at https://github.com/BioMedIA-MBZUAI/DGM-DR
Surgical tool classification and localization: results and methods from the MICCAI 2022 SurgToolLoc challenge
May 11, 2023



Abstract:The ability to automatically detect and track surgical instruments in endoscopic videos can enable transformational interventions. Assessing surgical performance and efficiency, identifying skilled tool use and choreography, and planning operational and logistical aspects of OR resources are just a few of the applications that could benefit. Unfortunately, obtaining the annotations needed to train machine learning models to identify and localize surgical tools is a difficult task. Annotating bounding boxes frame-by-frame is tedious and time-consuming, yet large amounts of data with a wide variety of surgical tools and surgeries must be captured for robust training. Moreover, ongoing annotator training is needed to stay up to date with surgical instrument innovation. In robotic-assisted surgery, however, potentially informative data like timestamps of instrument installation and removal can be programmatically harvested. The ability to rely on tool installation data alone would significantly reduce the workload to train robust tool-tracking models. With this motivation in mind we invited the surgical data science community to participate in the challenge, SurgToolLoc 2022. The goal was to leverage tool presence data as weak labels for machine learning models trained to detect tools and localize them in video frames with bounding boxes. We present the results of this challenge along with many of the team's efforts. We conclude by discussing these results in the broader context of machine learning and surgical data science. The training data used for this challenge consisting of 24,695 video clips with tool presence labels is also being released publicly and can be accessed at https://console.cloud.google.com/storage/browser/isi-surgtoolloc-2022.
RoboKit-MV: an Educational Initiative
Nov 22, 2021
Abstract:In this paper, we present a robot model and code base for affordable education in the field of humanoid robotics. We give an overview of the software and hardware of a robot that won several competitions with the team RoboKit in 2019-2021, provide analysis of the contemporary market of education in robotics, and highlight the reasoning beyond certain design solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge