Sharon Chokuwa
Divergent Domains, Convergent Grading: Enhancing Generalization in Diabetic Retinopathy Grading
Nov 04, 2024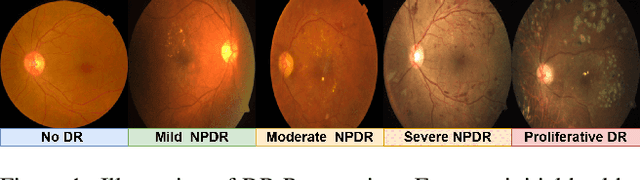
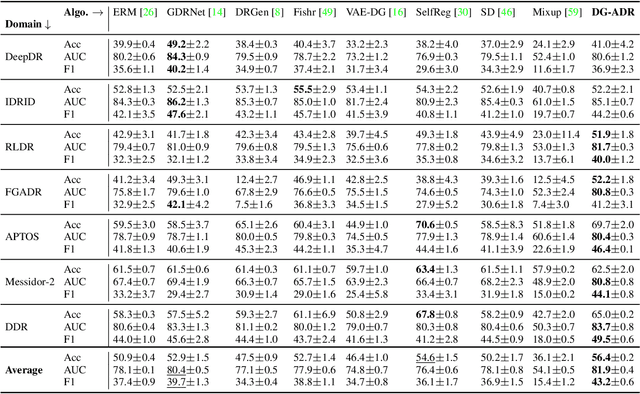
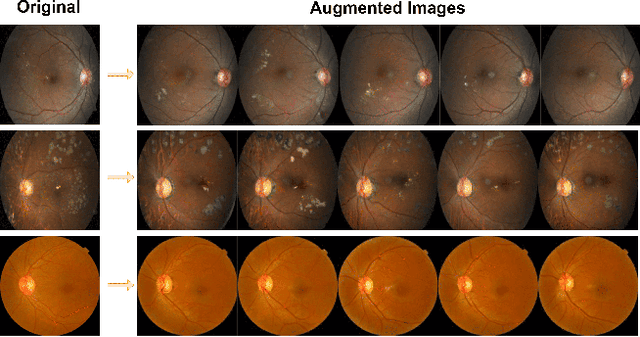

Abstract:Diabetic Retinopathy (DR) constitutes 5% of global blindness cases. While numerous deep learning approaches have sought to enhance traditional DR grading methods, they often falter when confronted with new out-of-distribution data thereby impeding their widespread application. In this study, we introduce a novel deep learning method for achieving domain generalization (DG) in DR grading and make the following contributions. First, we propose a new way of generating image-to-image diagnostically relevant fundus augmentations conditioned on the grade of the original fundus image. These augmentations are tailored to emulate the types of shifts in DR datasets thus increase the model's robustness. Second, we address the limitations of the standard classification loss in DG for DR fundus datasets by proposing a new DG-specific loss, domain alignment loss; which ensures that the feature vectors from all domains corresponding to the same class converge onto the same manifold for better domain generalization. Third, we tackle the coupled problem of data imbalance across DR domains and classes by proposing to employ Focal loss which seamlessly integrates with our new alignment loss. Fourth, due to inevitable observer variability in DR diagnosis that induces label noise, we propose leveraging self-supervised pretraining. This approach ensures that our DG model remains robust against early susceptibility to label noise, even when only a limited dataset of non-DR fundus images is available for pretraining. Our method demonstrates significant improvements over the strong Empirical Risk Minimization baseline and other recently proposed state-of-the-art DG methods for DR grading. Code is available at https://github.com/sharonchokuwa/dg-adr.
Generalizing Across Domains in Diabetic Retinopathy via Variational Autoencoders
Sep 20, 2023



Abstract:Domain generalization for Diabetic Retinopathy (DR) classification allows a model to adeptly classify retinal images from previously unseen domains with various imaging conditions and patient demographics, thereby enhancing its applicability in a wide range of clinical environments. In this study, we explore the inherent capacity of variational autoencoders to disentangle the latent space of fundus images, with an aim to obtain a more robust and adaptable domain-invariant representation that effectively tackles the domain shift encountered in DR datasets. Despite the simplicity of our approach, we explore the efficacy of this classical method and demonstrate its ability to outperform contemporary state-of-the-art approaches for this task using publicly available datasets. Our findings challenge the prevailing assumption that highly sophisticated methods for DR classification are inherently superior for domain generalization. This highlights the importance of considering simple methods and adapting them to the challenging task of generalizing medical images, rather than solely relying on advanced techniques.
DGM-DR: Domain Generalization with Mutual Information Regularized Diabetic Retinopathy Classification
Sep 18, 2023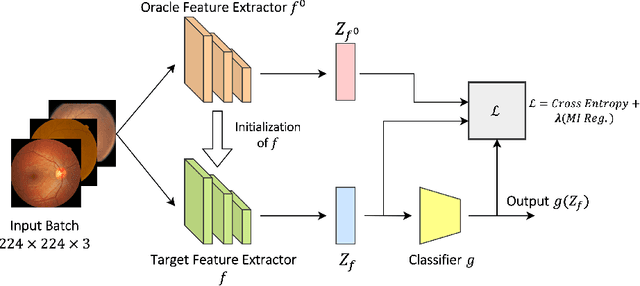
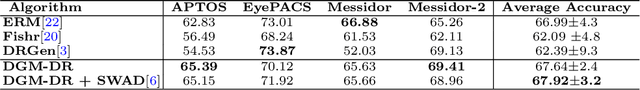
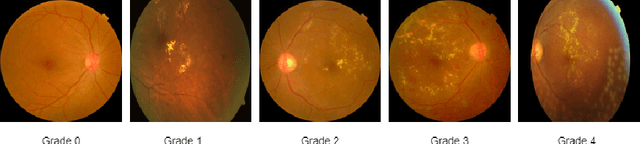
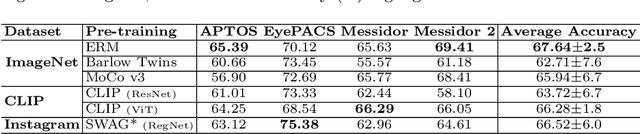
Abstract:The domain shift between training and testing data presents a significant challenge for training generalizable deep learning models. As a consequence, the performance of models trained with the independent and identically distributed (i.i.d) assumption deteriorates when deployed in the real world. This problem is exacerbated in the medical imaging context due to variations in data acquisition across clinical centers, medical apparatus, and patients. Domain generalization (DG) aims to address this problem by learning a model that generalizes well to any unseen target domain. Many domain generalization techniques were unsuccessful in learning domain-invariant representations due to the large domain shift. Furthermore, multiple tasks in medical imaging are not yet extensively studied in existing literature when it comes to DG point of view. In this paper, we introduce a DG method that re-establishes the model objective function as a maximization of mutual information with a large pretrained model to the medical imaging field. We re-visit the problem of DG in Diabetic Retinopathy (DR) classification to establish a clear benchmark with a correct model selection strategy and to achieve robust domain-invariant representation for an improved generalization. Moreover, we conduct extensive experiments on public datasets to show that our proposed method consistently outperforms the previous state-of-the-art by a margin of 5.25% in average accuracy and a lower standard deviation. Source code available at https://github.com/BioMedIA-MBZUAI/DGM-DR
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge