Akshay Goel
Health AI Developer Foundations
Nov 26, 2024



Abstract:Robust medical Machine Learning (ML) models have the potential to revolutionize healthcare by accelerating clinical research, improving workflows and outcomes, and producing novel insights or capabilities. Developing such ML models from scratch is cost prohibitive and requires substantial compute, data, and time (e.g., expert labeling). To address these challenges, we introduce Health AI Developer Foundations (HAI-DEF), a suite of pre-trained, domain-specific foundation models, tools, and recipes to accelerate building ML for health applications. The models cover various modalities and domains, including radiology (X-rays and computed tomography), histopathology, dermatological imaging, and audio. These models provide domain specific embeddings that facilitate AI development with less labeled data, shorter training times, and reduced computational costs compared to traditional approaches. In addition, we utilize a common interface and style across these models, and prioritize usability to enable developers to integrate HAI-DEF efficiently. We present model evaluations across various tasks and conclude with a discussion of their application and evaluation, covering the importance of ensuring efficacy, fairness, and equity. Finally, while HAI-DEF and specifically the foundation models lower the barrier to entry for ML in healthcare, we emphasize the importance of validation with problem- and population-specific data for each desired usage setting. This technical report will be updated over time as more modalities and features are added.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024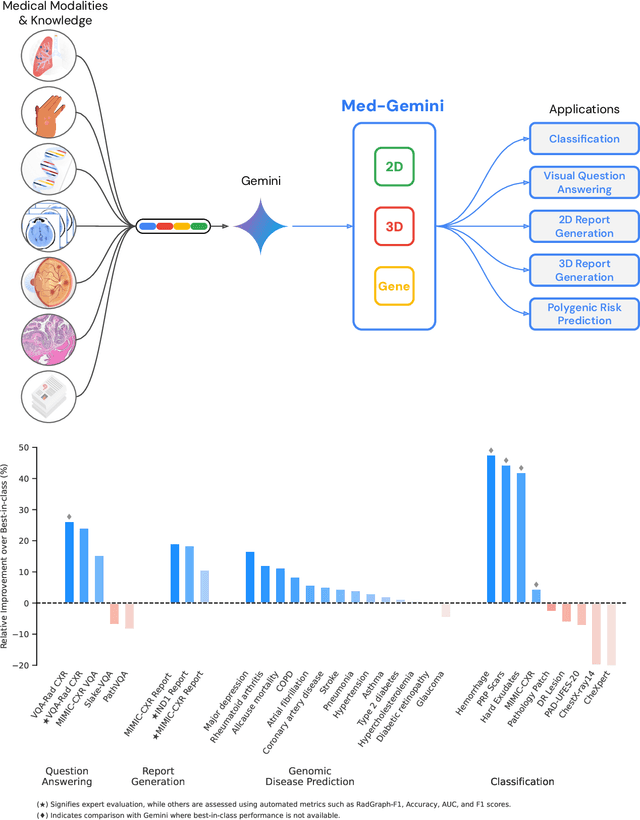
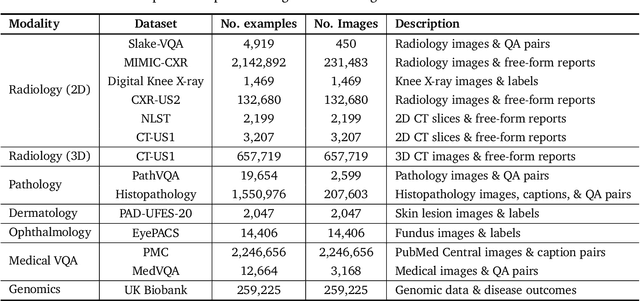

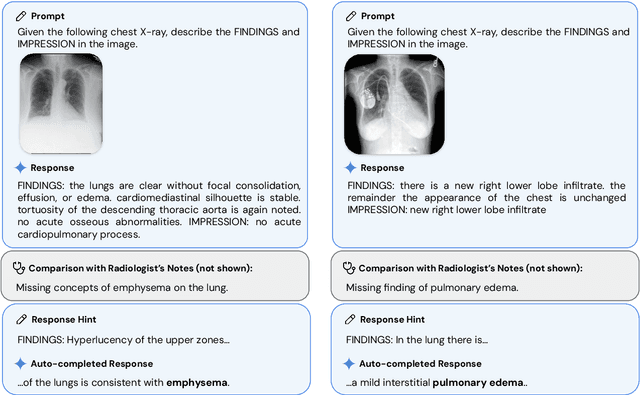
Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
LLMs Accelerate Annotation for Medical Information Extraction
Dec 04, 2023



Abstract:The unstructured nature of clinical notes within electronic health records often conceals vital patient-related information, making it challenging to access or interpret. To uncover this hidden information, specialized Natural Language Processing (NLP) models are required. However, training these models necessitates large amounts of labeled data, a process that is both time-consuming and costly when relying solely on human experts for annotation. In this paper, we propose an approach that combines Large Language Models (LLMs) with human expertise to create an efficient method for generating ground truth labels for medical text annotation. By utilizing LLMs in conjunction with human annotators, we significantly reduce the human annotation burden, enabling the rapid creation of labeled datasets. We rigorously evaluate our method on a medical information extraction task, demonstrating that our approach not only substantially cuts down on human intervention but also maintains high accuracy. The results highlight the potential of using LLMs to improve the utilization of unstructured clinical data, allowing for the swift deployment of tailored NLP solutions in healthcare.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge