Adi Szeskin
Simultaneous column-based deep learning progression analysis of atrophy associated with AMD in longitudinal OCT studies
Jul 31, 2023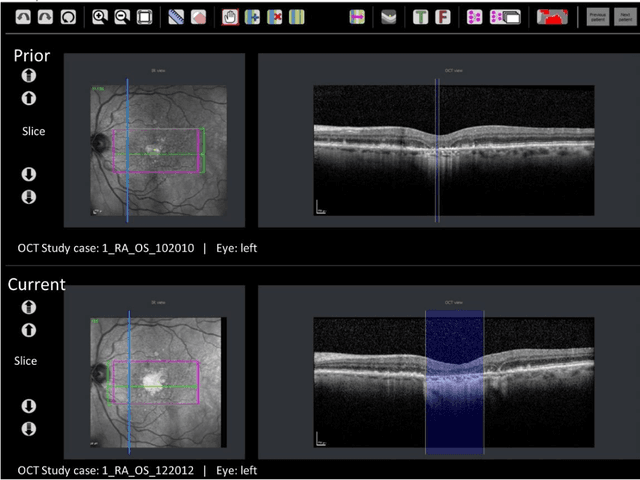
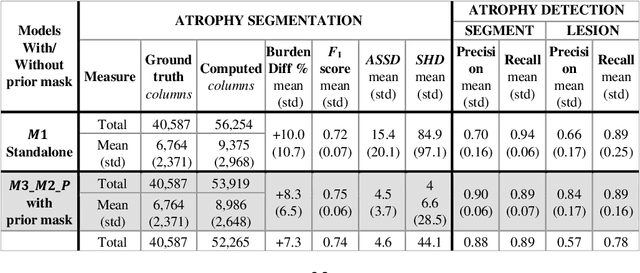
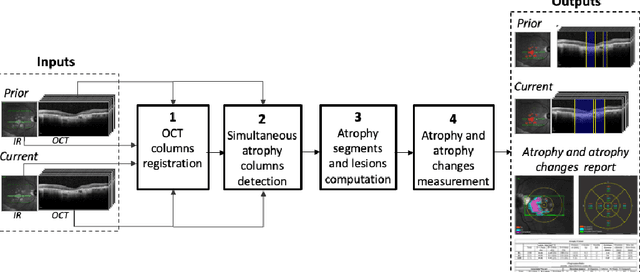
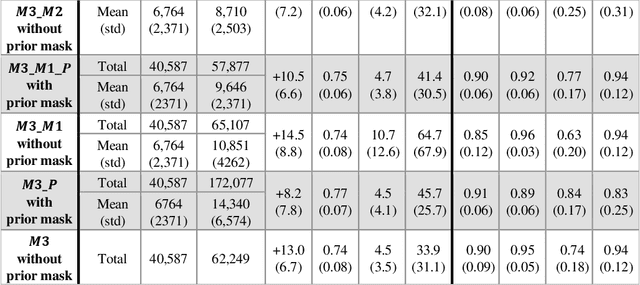
Abstract:Purpose: Disease progression of retinal atrophy associated with AMD requires the accurate quantification of the retinal atrophy changes on longitudinal OCT studies. It is based on finding, comparing, and delineating subtle atrophy changes on consecutive pairs (prior and current) of unregistered OCT scans. Methods: We present a fully automatic end-to-end pipeline for the simultaneous detection and quantification of time-related atrophy changes associated with dry AMD in pairs of OCT scans of a patient. It uses a novel simultaneous multi-channel column-based deep learning model trained on registered pairs of OCT scans that concurrently detects and segments retinal atrophy segments in consecutive OCT scans by classifying light scattering patterns in matched pairs of vertical pixel-wide columns (A-scans) in registered prior and current OCT slices (B-scans). Results: Experimental results on 4,040 OCT slices with 5.2M columns from 40 scans pairs of 18 patients (66% training/validation, 33% testing) with 24.13+-14.0 months apart in which Complete RPE and Outer Retinal Atrophy (cRORA) was identified in 1,998 OCT slices (735 atrophy lesions from 3,732 segments, 0.45M columns) yield a mean atrophy segments detection precision, recall of 0.90+-0.09, 0.95+-0.06 and 0.74+-0.18, 0.94+-0.12 for atrophy lesions with AUC=0.897, all above observer variability. Simultaneous classification outperforms standalone classification precision and recall by 30+-62% and 27+-0% for atrophy segments and lesions. Conclusions: simultaneous column-based detection and quantification of retinal atrophy changes associated with AMD is accurate and outperforms standalone classification methods. Translational relevance: an automatic and efficient way to detect and quantify retinal atrophy changes associated with AMD.
A Weak Supervision Approach to Detecting Visual Anomalies for Automated Testing of Graphics Units
Dec 09, 2019

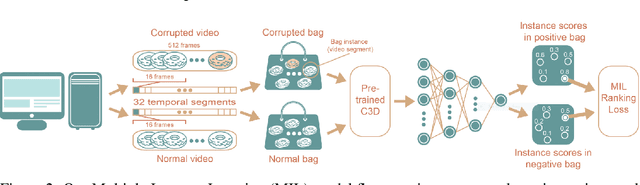
Abstract:We present a deep learning system for testing graphics units by detecting novel visual corruptions in videos. Unlike previous work in which manual tagging was required to collect labeled training data, our weak supervision method is fully automatic and needs no human labelling. This is achieved by reproducing driver bugs that increase the probability of generating corruptions, and by making use of ideas and methods from the Multiple Instance Learning (MIL) setting. In our experiments, we significantly outperform unsupervised methods such as GAN-based models and discover novel corruptions undetected by baselines, while adhering to strict requirements on accuracy and efficiency of our real-time system.
Automatic detection and diagnosis of sacroiliitis in CT scans as incidental findings
Aug 14, 2019



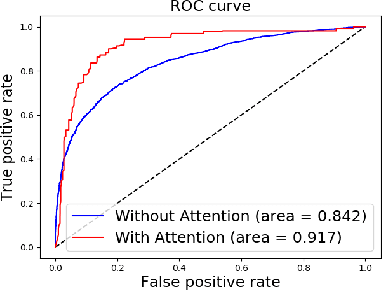
Abstract:Early diagnosis of sacroiliitis may lead to preventive treatment which can significantly improve the patient's quality of life in the long run. Oftentimes, a CT scan of the lower back or abdomen is acquired for suspected back pain. However, since the differences between a healthy and an inflamed sacroiliac joint in the early stages are subtle, the condition may be missed. We have developed a new automatic algorithm for the diagnosis and grading of sacroiliitis CT scans as incidental findings, for patients who underwent CT scanning as part of their lower back pain workout. The method is based on supervised machine and deep learning techniques. The input is a CT scan that includes the patient's pelvis. The output is a diagnosis for each sacroiliac joint. The algorithm consists of four steps: 1) computation of an initial region of interest (ROI) that includes the pelvic joints region using heuristics and a U-Net classifier; 2) refinement of the ROI to detect both sacroiliac joints using a four-tree random forest; 3) individual sacroiliitis grading of each sacroiliac joint in each CT slice with a custom slice CNN classifier, and; 4) sacroiliitis diagnosis and grading by combining the individual slice grades using a random forest. Experimental results on 484 sacroiliac joints yield a binary and a 3-class case classification accuracy of 91.9% and 86%, a sensitivity of 95% and 82%, and an Area-Under-the-Curve of 0.97 and 0.57, respectively. Automatic computer-based analysis of CT scans has the potential of being a useful method for the diagnosis and grading of sacroiliitis as an incidental finding.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019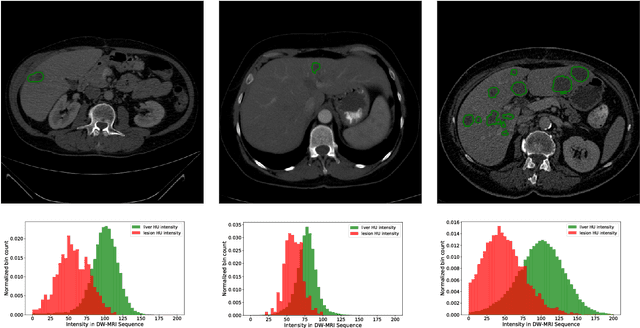
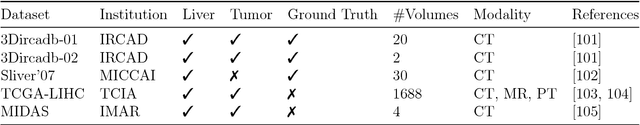
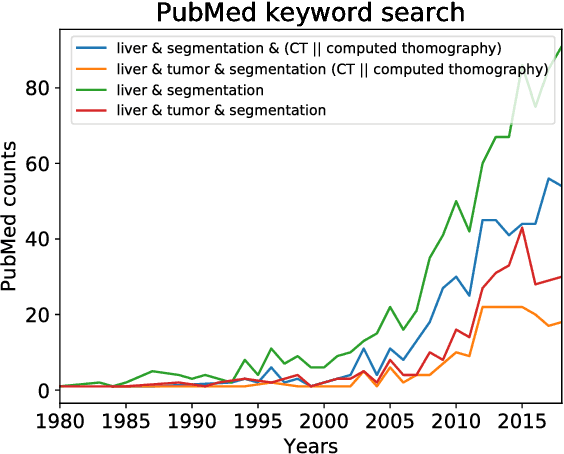
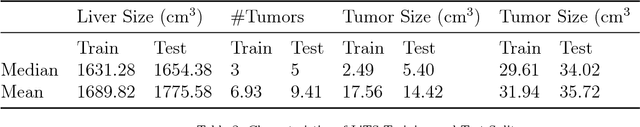
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge