Zhenchang Wang
Image Reconstruction for Accelerated MR Scan with Faster Fourier Convolutional Neural Networks
Jun 05, 2023



Abstract:Partial scan is a common approach to accelerate Magnetic Resonance Imaging (MRI) data acquisition in both 2D and 3D settings. However, accurately reconstructing images from partial scan data (i.e., incomplete k-space matrices) remains challenging due to lack of an effectively global receptive field in both spatial and k-space domains. To address this problem, we propose the following: (1) a novel convolutional operator called Faster Fourier Convolution (FasterFC) to replace the two consecutive convolution operations typically used in convolutional neural networks (e.g., U-Net, ResNet). Based on the spectral convolution theorem in Fourier theory, FasterFC employs alternating kernels of size 1 in 3D case) in different domains to extend the dual-domain receptive field to the global and achieves faster calculation speed than traditional Fast Fourier Convolution (FFC). (2) A 2D accelerated MRI method, FasterFC-End-to-End-VarNet, which uses FasterFC to improve the sensitivity maps and reconstruction quality. (3) A multi-stage 3D accelerated MRI method called FasterFC-based Single-to-group Network (FAS-Net) that utilizes a single-to-group algorithm to guide k-space domain reconstruction, followed by FasterFC-based cascaded convolutional neural networks to expand the effective receptive field in the dual-domain. Experimental results on the fastMRI and Stanford MRI Data datasets demonstrate that FasterFC improves the quality of both 2D and 3D reconstruction. Moreover, FAS-Net, as a 3D high-resolution multi-coil (eight) accelerated MRI method, achieves superior reconstruction performance in both qualitative and quantitative results compared with state-of-the-art 2D and 3D methods.
Dual-Domain Reconstruction Networks with V-Net and K-Net for fast MRI
Mar 14, 2022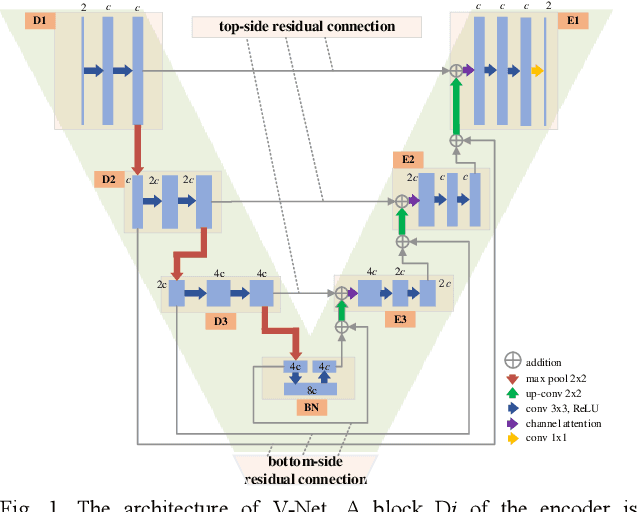
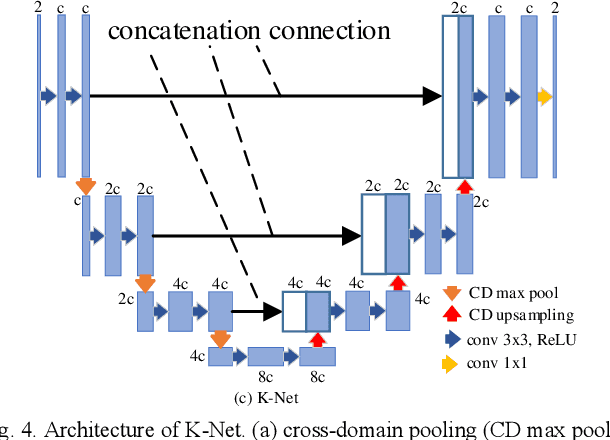
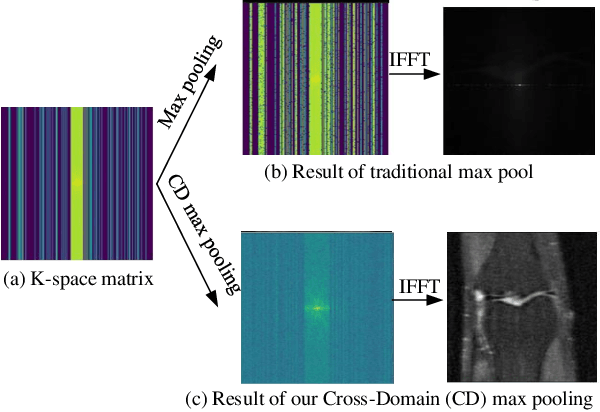
Abstract:Purpose: To introduce a dual-domain reconstruction network with V-Net and K-Net for accurate MR image reconstruction from undersampled k-space data. Methods: Most state-of-the-art reconstruction methods apply U-Net or cascaded U-Nets in image domain and/or k-space domain. Nevertheless, these methods have following problems: (1) Directly applying U-Net in k-space domain is not optimal for extracting features in k-space domain; (2) Classical image-domain oriented U-Net is heavy-weight and hence is inefficient to be cascaded many times for yielding good reconstruction accuracy; (3) Classical image-domain oriented U-Net does not fully make use information of encoder network for extracting features in decoder network; and (4) Existing methods are ineffective in simultaneously extracting and fusing features in image domain and its dual k-space domain. To tackle these problems, we propose in this paper (1) an image-domain encoder-decoder sub-network called V-Net which is more light-weight for cascading and effective in fully utilizing features in the encoder for decoding, (2) a k-space domain sub-network called K-Net which is more suitable for extracting hierarchical features in k-space domain, and (3) a dual-domain reconstruction network where V-Nets and K-Nets are parallelly and effectively combined and cascaded. Results: Extensive experimental results on the challenging fastMRI dataset demonstrate that the proposed KV-Net can reconstruct high-quality images and outperform current state-of-the-art approaches with fewer parameters. Conclusions: To reconstruct images effectively and efficiently from incomplete k-space data, we have presented a parallel dual-domain KV-Net to combine K-Nets and V-Nets. The KV-Net is more lightweight than state-of-the-art methods but achieves better reconstruction performance.
Active Phase-Encode Selection for Slice-Specific Fast MR Scanning Using a Transformer-Based Deep Reinforcement Learning Framework
Mar 11, 2022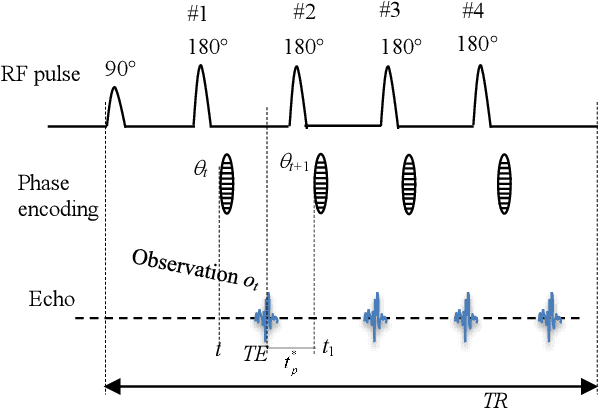
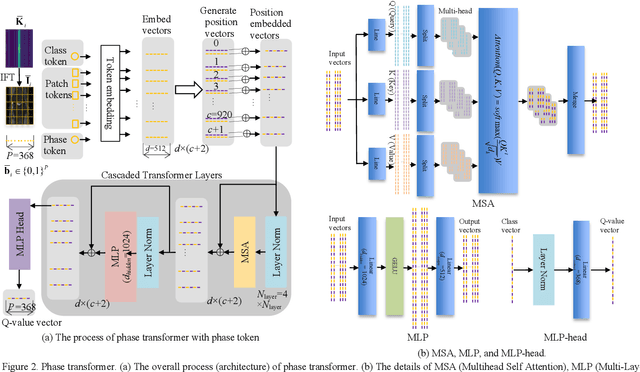
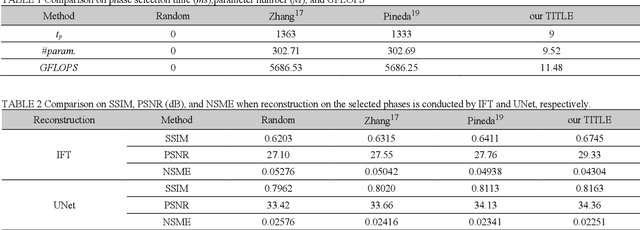
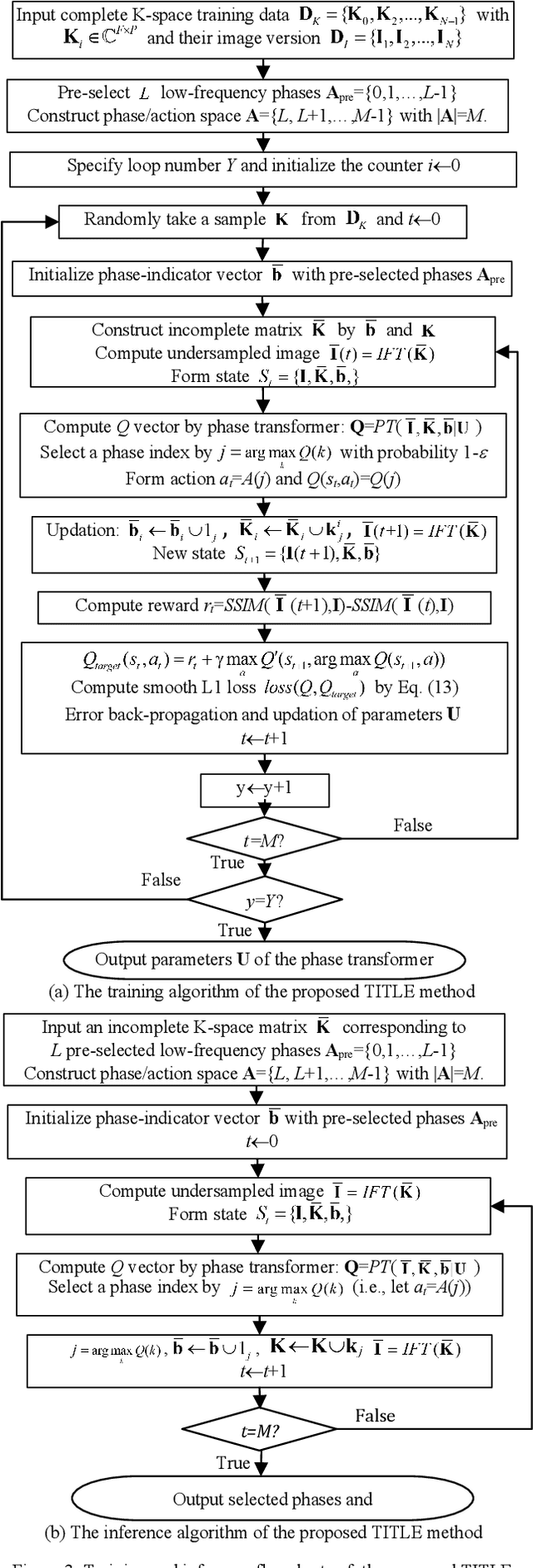
Abstract:Purpose: Long scan time in phase encoding for forming complete K-space matrices is a critical drawback of MRI, making patients uncomfortable and wasting important time for diagnosing emergent diseases. This paper aims to reducing the scan time by actively and sequentially selecting partial phases in a short time so that a slice can be accurately reconstructed from the resultant slice-specific incomplete K-space matrix. Methods: A transformer based deep reinforcement learning framework is proposed for actively determining a sequence of partial phases according to reconstruction-quality based Q-value (a function of reward), where the reward is the improvement degree of reconstructed image quality. The Q-value is efficiently predicted from binary phase-indicator vectors, incomplete K-space matrices and their corresponding undersampled images with a light-weight transformer so that the sequential information of phases and global relationship in images can be used. The inverse Fourier transform is employed for efficiently computing the undersampled images and hence gaining the rewards of selecting phases. Results: Experimental results on the fastMRI dataset with original K-space data accessible demonstrate the efficiency and accuracy superiorities of proposed method. Compared with the state-of-the-art reinforcement learning based method proposed by Pineda et al., the proposed method is roughly 150 times faster and achieves significant improvement in reconstruction accuracy. Conclusions: We have proposed a light-weight transformer based deep reinforcement learning framework for generating high-quality slice-specific trajectory consisting of a small number of phases. The proposed method, called TITLE (Transformer Involved Trajectory LEarning), has remarkable superiority in phase-encode selection efficiency and image reconstruction accuracy.
Ricci Curvature Based Volumetric Segmentation of the Auditory Ossicles
Jun 26, 2020
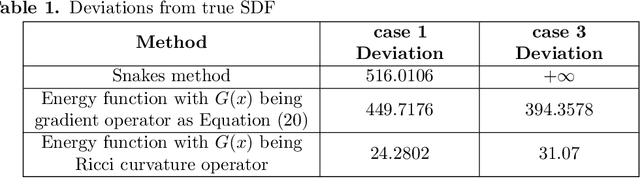

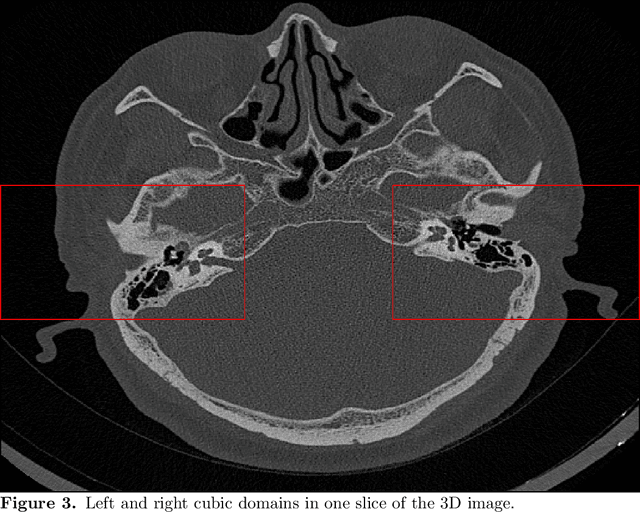
Abstract:The auditory ossicles that are located in the middle ear are the smallest bones in the human body. Their damage will result in hearing loss. It is therefore important to be able to automatically diagnose ossicles' diseases based on Computed Tomography (CT) 3D imaging. However CT images usually include the whole head area, which is much larger than the bones of interest, thus the localization of the ossicles, followed by segmentation, both play a significant role in automatic diagnosis. The commonly employed local segmentation methods require manually selected initial points, which is a highly time consuming process. We therefore propose a completely automatic method to locate the ossicles which requires neither templates, nor manual labels. It relies solely on the connective properties of the auditory ossicles themselves, and their relationship with the surrounding tissue fluid. For the segmentation task, we define a novel energy function and obtain the shape of the ossicles from the 3D CT image by minimizing this new energy. Compared to the state-of-the-art methods which usually use the gradient operator and some normalization terms, we propose to add a Ricci curvature term to the commonly employed energy function. We compare our proposed method with the state-of-the-art methods and show that the performance of discrete Forman-Ricci curvature is superior to the others.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge