Zeya Wang
Towards Proactive Personalization through Profile Customization for Individual Users in Dialogues
Dec 17, 2025Abstract:The deployment of Large Language Models (LLMs) in interactive systems necessitates a deep alignment with the nuanced and dynamic preferences of individual users. Current alignment techniques predominantly address universal human values or static, single-turn preferences, thereby failing to address the critical needs of long-term personalization and the initial user cold-start problem. To bridge this gap, we propose PersonalAgent, a novel user-centric lifelong agent designed to continuously infer and adapt to user preferences. PersonalAgent constructs and dynamically refines a unified user profile by decomposing dialogues into single-turn interactions, framing preference inference as a sequential decision-making task. Experiments show that PersonalAgent achieves superior performance over strong prompt-based and policy optimization baselines, not only in idealized but also in noisy conversational contexts, while preserving cross-session preference consistency. Furthermore, human evaluation confirms that PersonalAgent excels at capturing user preferences naturally and coherently. Our findings underscore the importance of lifelong personalization for developing more inclusive and adaptive conversational agents. Our code is available here.
Deep Clustering Evaluation: How to Validate Internal Clustering Validation Measures
Mar 21, 2024Abstract:Deep clustering, a method for partitioning complex, high-dimensional data using deep neural networks, presents unique evaluation challenges. Traditional clustering validation measures, designed for low-dimensional spaces, are problematic for deep clustering, which involves projecting data into lower-dimensional embeddings before partitioning. Two key issues are identified: 1) the curse of dimensionality when applying these measures to raw data, and 2) the unreliable comparison of clustering results across different embedding spaces stemming from variations in training procedures and parameter settings in different clustering models. This paper addresses these challenges in evaluating clustering quality in deep learning. We present a theoretical framework to highlight ineffectiveness arising from using internal validation measures on raw and embedded data and propose a systematic approach to applying clustering validity indices in deep clustering contexts. Experiments show that this framework aligns better with external validation measures, effectively reducing the misguidance from the improper use of clustering validity indices in deep learning.
SODA: Detecting Covid-19 in Chest X-rays with Semi-supervised Open Set Domain Adaptation
May 22, 2020
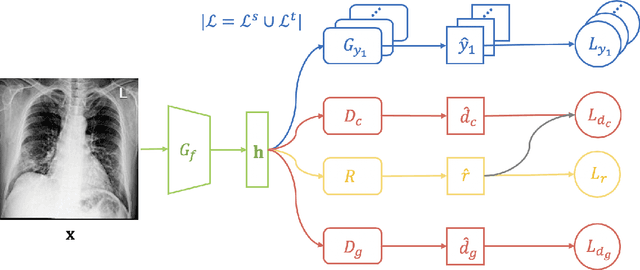
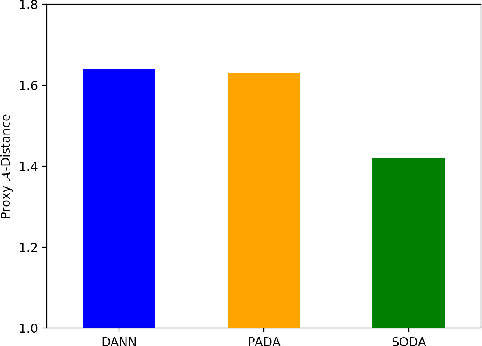
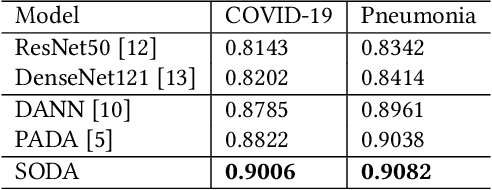
Abstract:The global pandemic of COVID-19 has infected millions of people since its first outbreak in last December. A key challenge for preventing and controlling COVID-19 is how to quickly, widely, and effectively implement the test for the disease, because testing is the first step to break the chains of transmission. To assist the diagnosis of the disease, radiology imaging is used to complement the screening process and triage patients into different risk levels. Deep learning methods have taken a more active role in automatically detecting COVID-19 disease in chest x-ray images, as witnessed in many recent works. Most of these works first train a CNN on an existing large-scale chest x-ray image dataset and then fine-tune it with a COVID-19 dataset at a much smaller scale. However, direct transfer across datasets from different domains may lead to poor performance due to visual domain shift. Also, the small scale of the COVID-19 dataset on the target domain can make the training fall into the overfitting trap. To solve all these crucial problems and fully exploit the available large-scale chest x-ray image dataset, we formulate the problem of COVID-19 chest x-ray image classification in a semi-supervised open set domain adaptation setting, through which we are motivated to reduce the domain shift and avoid overfitting when training on a very small dataset of COVID-19. In addressing this formulated problem, we propose a novel Semi-supervised Open set Domain Adversarial network (SODA), which is able to align the data distributions across different domains in a general domain space and also in a common subspace of source and target data. In our experiments, SODA achieves a leading classification performance compared with recent state-of-the-art models, as well as effectively separating COVID-19 with common pneumonia.
Show, Describe and Conclude: On Exploiting the Structure Information of Chest X-Ray Reports
Apr 26, 2020
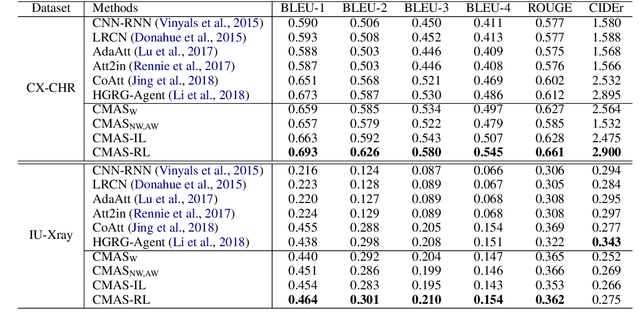

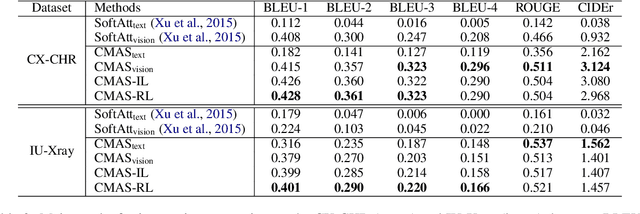
Abstract:Chest X-Ray (CXR) images are commonly used for clinical screening and diagnosis. Automatically writing reports for these images can considerably lighten the workload of radiologists for summarizing descriptive findings and conclusive impressions. The complex structures between and within sections of the reports pose a great challenge to the automatic report generation. Specifically, the section Impression is a diagnostic summarization over the section Findings; and the appearance of normality dominates each section over that of abnormality. Existing studies rarely explore and consider this fundamental structure information. In this work, we propose a novel framework that exploits the structure information between and within report sections for generating CXR imaging reports. First, we propose a two-stage strategy that explicitly models the relationship between Findings and Impression. Second, we design a novel cooperative multi-agent system that implicitly captures the imbalanced distribution between abnormality and normality. Experiments on two CXR report datasets show that our method achieves state-of-the-art performance in terms of various evaluation metrics. Our results expose that the proposed approach is able to generate high-quality medical reports through integrating the structure information.
Adversarial Domain Adaptation Being Aware of Class Relationships
May 28, 2019

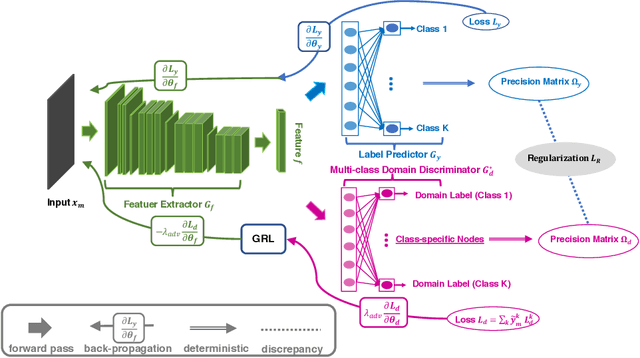
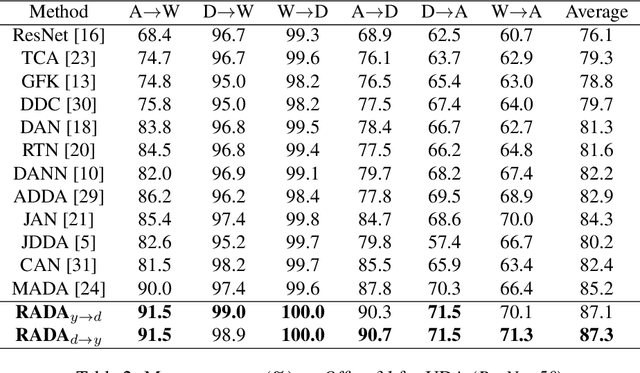
Abstract:Adversarial training is a useful approach to promote the learning of transferable representations across the source and target domains, which has been widely applied for domain adaptation (DA) tasks based on deep neural networks. Until very recently, existing adversarial domain adaptation (ADA) methods ignore the useful information from the label space, which is an important factor accountable for the complicated data distributions associated with different semantic classes. Especially, the inter-class semantic relationships have been rarely considered and discussed in the current work of transfer learning. In this paper, we propose a novel relationship-aware adversarial domain adaptation (RADA) algorithm, which first utilizes a single multi-class domain discriminator to enforce the learning of inter-class dependency structure during domain-adversarial training and then aligns this structure with the inter-class dependencies that are characterized from training the label predictor on the source domain. Specifically, we impose a regularization term to penalize the structure discrepancy between the inter-class dependencies respectively estimated from domain discriminator and label predictor. Through this alignment, our proposed method makes the ADA aware of class relationships. Empirical studies show that the incorporation of class relationships significantly improves the performance on benchmark datasets.
Reinforced Auto-Zoom Net: Towards Accurate and Fast Breast Cancer Segmentation in Whole-slide Images
Jul 29, 2018
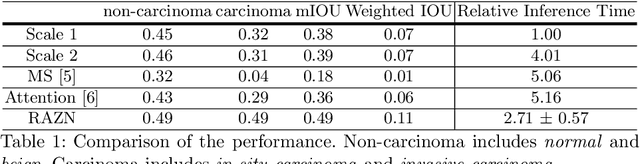

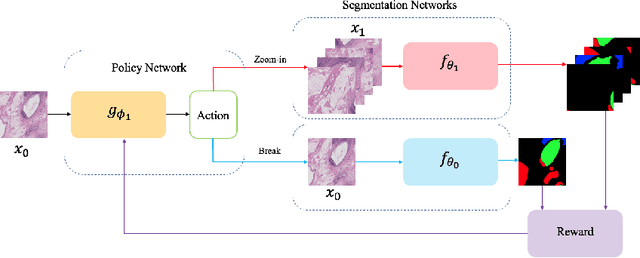
Abstract:Convolutional neural networks have led to significant breakthroughs in the domain of medical image analysis. However, the task of breast cancer segmentation in whole-slide images (WSIs) is still underexplored. WSIs are large histopathological images with extremely high resolution. Constrained by the hardware and field of view, using high-magnification patches can slow down the inference process and using low-magnification patches can cause the loss of information. In this paper, we aim to achieve two seemingly conflicting goals for breast cancer segmentation: accurate and fast prediction. We propose a simple yet efficient framework Reinforced Auto-Zoom Net (RAZN) to tackle this task. Motivated by the zoom-in operation of a pathologist using a digital microscope, RAZN learns a policy network to decide whether zooming is required in a given region of interest. Because the zoom-in action is selective, RAZN is robust to unbalanced and noisy ground truth labels and can efficiently reduce overfitting. We evaluate our method on a public breast cancer dataset. RAZN outperforms both single-scale and multi-scale baseline approaches, achieving better accuracy at low inference cost.
Unsupervised Domain Adaptation for Automatic Estimation of Cardiothoracic Ratio
Jul 10, 2018
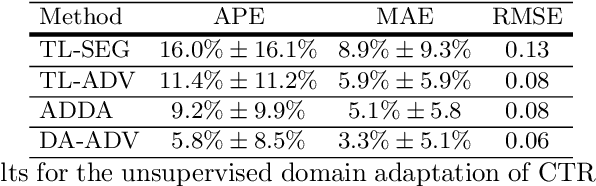


Abstract:The cardiothoracic ratio (CTR), a clinical metric of heart size in chest X-rays (CXRs), is a key indicator of cardiomegaly. Manual measurement of CTR is time-consuming and can be affected by human subjectivity, making it desirable to design computer-aided systems that assist clinicians in the diagnosis process. Automatic CTR estimation through chest organ segmentation, however, requires large amounts of pixel-level annotated data, which is often unavailable. To alleviate this problem, we propose an unsupervised domain adaptation framework based on adversarial networks. The framework learns domain invariant feature representations from openly available data sources to produce accurate chest organ segmentation for unlabeled datasets. Specifically, we propose a model that enforces our intuition that prediction masks should be domain independent. Hence, we introduce a discriminator that distinguishes segmentation predictions from ground truth masks. We evaluate our system's prediction based on the assessment of radiologists and demonstrate the clinical practicability for the diagnosis of cardiomegaly. We finally illustrate on the JSRT dataset that the semi-supervised performance of our model is also very promising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge