Yijun Zhou
Static Is Not Enough: A Comparative Study of VR and SpaceMouse in Static and Dynamic Teleoperation Tasks
Jan 19, 2026Abstract:Imitation learning relies on high-quality demonstrations, and teleoperation is a primary way to collect them, making teleoperation interface choice crucial for the data. Prior work mainly focused on static tasks, i.e., discrete, segmented motions, yet demonstrations also include dynamic tasks requiring reactive control. As dynamic tasks impose fundamentally different interface demands, insights from static-task evaluations cannot generalize. To address this gap, we conduct a within-subjects study comparing a VR controller and a SpaceMouse across two static and two dynamic tasks ($N=25$). We assess success rate, task duration, cumulative success, alongside NASA-TLX, SUS, and open-ended feedback. Results show statistically significant advantages for VR: higher success rates, particularly on dynamic tasks, shorter successful execution times across tasks, and earlier successes across attempts, with significantly lower workload and higher usability. As existing VR teleoperation systems are rarely open-source or suited for dynamic tasks, we release our VR interface to fill this gap.
Multimodal Feature Fusion Network with Text Difference Enhancement for Remote Sensing Change Detection
Sep 04, 2025Abstract:Although deep learning has advanced remote sensing change detection (RSCD), most methods rely solely on image modality, limiting feature representation, change pattern modeling, and generalization especially under illumination and noise disturbances. To address this, we propose MMChange, a multimodal RSCD method that combines image and text modalities to enhance accuracy and robustness. An Image Feature Refinement (IFR) module is introduced to highlight key regions and suppress environmental noise. To overcome the semantic limitations of image features, we employ a vision language model (VLM) to generate semantic descriptions of bitemporal images. A Textual Difference Enhancement (TDE) module then captures fine grained semantic shifts, guiding the model toward meaningful changes. To bridge the heterogeneity between modalities, we design an Image Text Feature Fusion (ITFF) module that enables deep cross modal integration. Extensive experiments on LEVIRCD, WHUCD, and SYSUCD demonstrate that MMChange consistently surpasses state of the art methods across multiple metrics, validating its effectiveness for multimodal RSCD. Code is available at: https://github.com/yikuizhai/MMChange.
scFusionTTT: Single-cell transcriptomics and proteomics fusion with Test-Time Training layers
Oct 17, 2024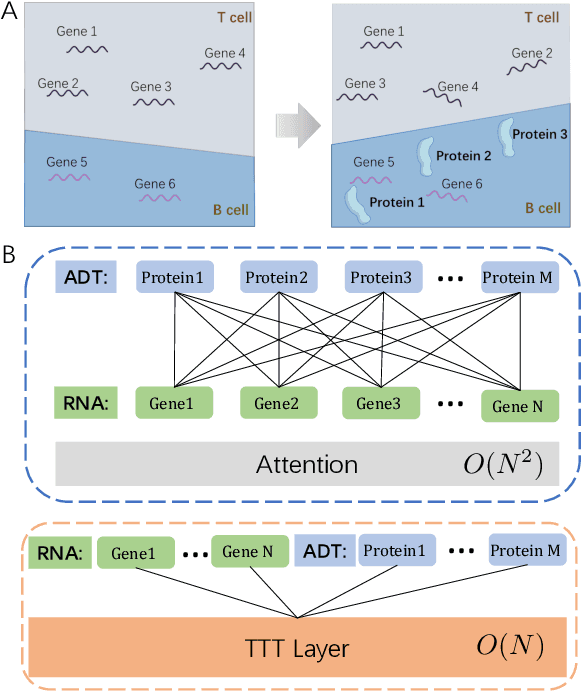
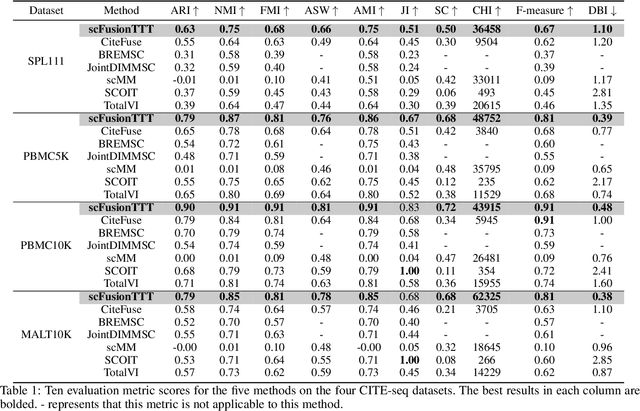
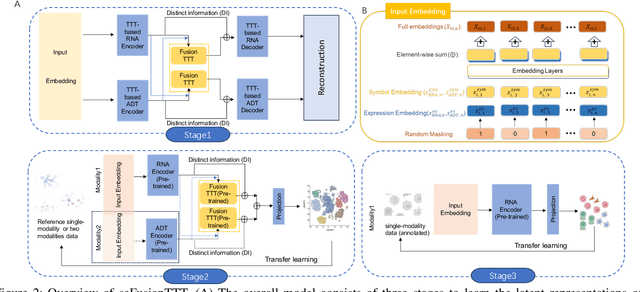
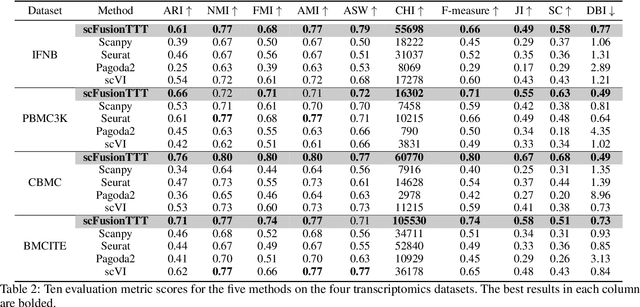
Abstract:Single-cell multi-omics (scMulti-omics) refers to the paired multimodal data, such as Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq), where the regulation of each cell was measured from different modalities, i.e. genes and proteins. scMulti-omics can reveal heterogeneity inside tumors and understand the distinct genetic properties of diverse cell types, which is crucial to targeted therapy. Currently, deep learning methods based on attention structures in the bioinformatics area face two challenges. The first challenge is the vast number of genes in a single cell. Traditional attention-based modules struggled to effectively leverage all gene information due to their limited capacity for long-context learning and high-complexity computing. The second challenge is that genes in the human genome are ordered and influence each other's expression. Most of the methods ignored this sequential information. The recently introduced Test-Time Training (TTT) layer is a novel sequence modeling approach, particularly suitable for handling long contexts like genomics data because TTT layer is a linear complexity sequence modeling structure and is better suited to data with sequential relationships. In this paper, we propose scFusionTTT, a novel method for Single-Cell multimodal omics Fusion with TTT-based masked autoencoder. Of note, we combine the order information of genes and proteins in the human genome with the TTT layer, fuse multimodal omics, and enhance unimodal omics analysis. Finally, the model employs a three-stage training strategy, which yielded the best performance across most metrics in four multimodal omics datasets and four unimodal omics datasets, demonstrating the superior performance of our model. The dataset and code will be available on https://github.com/DM0815/scFusionTTT.
Coalitions of AI-based Methods Predict 15-Year Risks of Breast Cancer Metastasis Using Real-World Clinical Data with AUC up to 0.9
Aug 29, 2024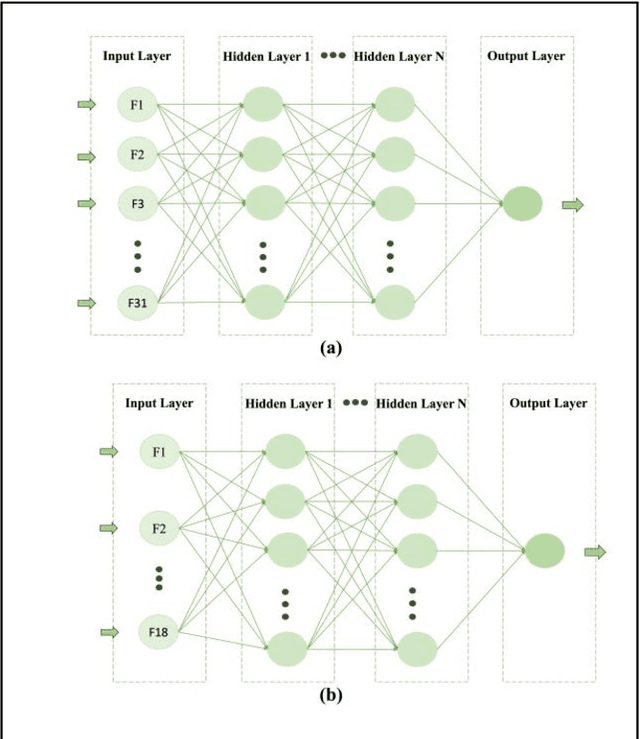
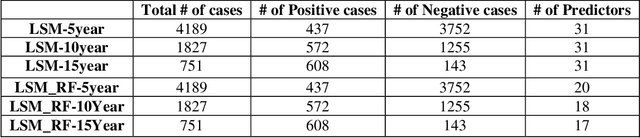
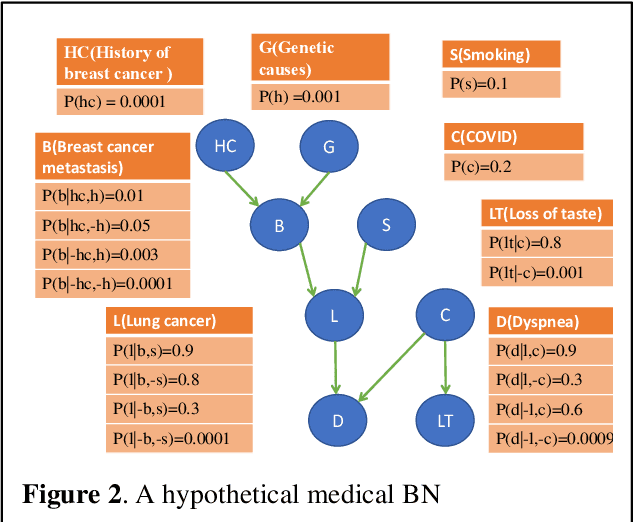
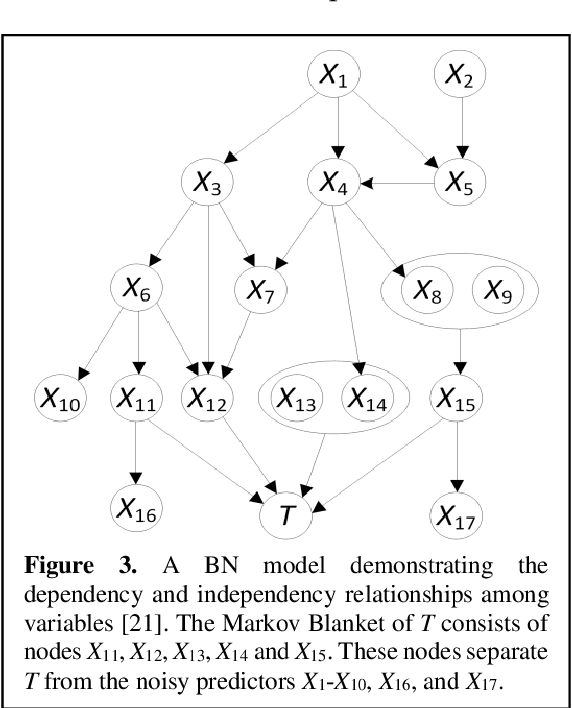
Abstract:Breast cancer is one of the two cancers responsible for the most deaths in women, with about 42,000 deaths each year in the US. That there are over 300,000 breast cancers newly diagnosed each year suggests that only a fraction of the cancers result in mortality. Thus, most of the women undergo seemingly curative treatment for localized cancers, but a significant later succumb to metastatic disease for which current treatments are only temporizing for the vast majority. The current prognostic metrics are of little actionable value for 4 of the 5 women seemingly cured after local treatment, and many women are exposed to morbid and even mortal adjuvant therapies unnecessarily, with these adjuvant therapies reducing metastatic recurrence by only a third. Thus, there is a need for better prognostics to target aggressive treatment at those who are likely to relapse and spare those who were actually cured. While there is a plethora of molecular and tumor-marker assays in use and under-development to detect recurrence early, these are time consuming, expensive and still often un-validated as to actionable prognostic utility. A different approach would use large data techniques to determine clinical and histopathological parameters that would provide accurate prognostics using existing data. Herein, we report on machine learning, together with grid search and Bayesian Networks to develop algorithms that present a AUC of up to 0.9 in ROC analyses, using only extant data. Such algorithms could be rapidly translated to clinical management as they do not require testing beyond routine tumor evaluations.
Deep Learning to Predict Late-Onset Breast Cancer Metastasis: the Single Hyperparameter Grid Search (SHGS) Strategy for Meta Tuning Concerning Deep Feed-forward Neural Network
Aug 28, 2024

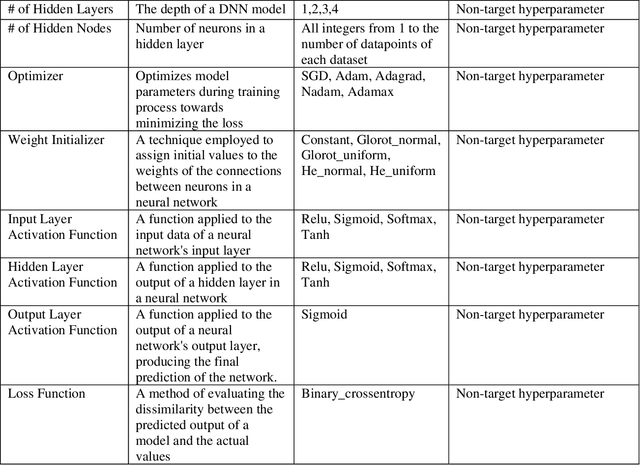
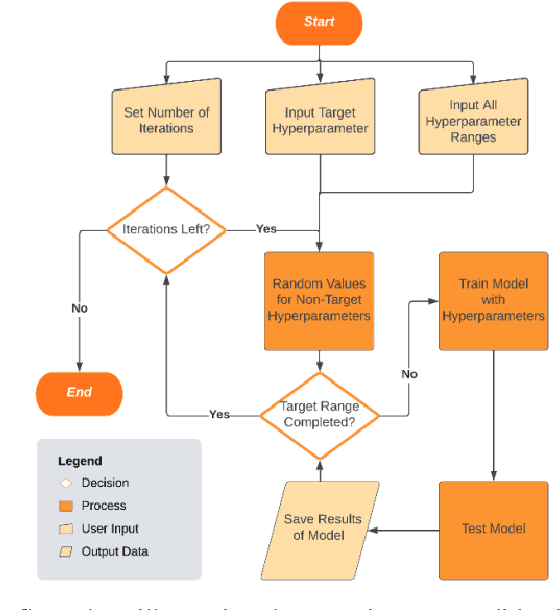
Abstract:While machine learning has advanced in medicine, its widespread use in clinical applications, especially in predicting breast cancer metastasis, is still limited. We have been dedicated to constructing a DFNN model to predict breast cancer metastasis n years in advance. However, the challenge lies in efficiently identifying optimal hyperparameter values through grid search, given the constraints of time and resources. Issues such as the infinite possibilities for continuous hyperparameters like l1 and l2, as well as the time-consuming and costly process, further complicate the task. To address these challenges, we developed Single Hyperparameter Grid Search (SHGS) strategy, serving as a preselection method before grid search. Our experiments with SHGS applied to DFNN models for breast cancer metastasis prediction focus on analyzing eight target hyperparameters: epochs, batch size, dropout, L1, L2, learning rate, decay, and momentum. We created three figures, each depicting the experiment results obtained from three LSM-I-10-Plus-year datasets. These figures illustrate the relationship between model performance and the target hyperparameter values. For each hyperparameter, we analyzed whether changes in this hyperparameter would affect model performance, examined if there were specific patterns, and explored how to choose values for the particular hyperparameter. Our experimental findings reveal that the optimal value of a hyperparameter is not only dependent on the dataset but is also significantly influenced by the settings of other hyperparameters. Additionally, our experiments suggested some reduced range of values for a target hyperparameter, which may be helpful for low-budget grid search. This approach serves as a prior experience and foundation for subsequent use of grid search to enhance model performance.
Deep Learning: a Heuristic Three-stage Mechanism for Grid Searches to Optimize the Future Risk Prediction of Breast Cancer Metastasis Using EHR-based Clinical Data
Aug 15, 2024Abstract:A grid search, at the cost of training and testing a large number of models, is an effective way to optimize the prediction performance of deep learning models. A challenging task concerning grid search is the time management. Without a good time management scheme, a grid search can easily be set off as a mission that will not finish in our lifetime. In this study, we introduce a heuristic three-stage mechanism for managing the running time of low-budget grid searches, and the sweet-spot grid search (SSGS) and randomized grid search (RGS) strategies for improving model prediction performance, in predicting the 5-year, 10-year, and 15-year risk of breast cancer metastasis. We develop deep feedforward neural network (DFNN) models and optimize them through grid searches. We conduct eight cycles of grid searches by applying our three-stage mechanism and SSGS and RGS strategies. We conduct various SHAP analyses including unique ones that interpret the importance of the DFNN-model hyperparameters. Our results show that grid search can greatly improve model prediction. The grid searches we conducted improved the risk prediction of 5-year, 10-year, and 15-year breast cancer metastasis by 18.6%, 16.3%, and 17.3% respectively, over the average performance of all corresponding models we trained using the RGS strategy. We not only demonstrate best model performance but also characterize grid searches from various aspects such as their capabilities of discovering decent models and the unit grid search time. The three-stage mechanism worked effectively. It made our low-budget grid searches feasible and manageable, and in the meantime helped improve model prediction performance. Our SHAP analyses identified both clinical risk factors important for the prediction of future risk of breast cancer metastasis, and DFNN-model hyperparameters important to the prediction of performance scores.
WHENet: Real-time Fine-Grained Estimation for Wide Range Head Pose
May 20, 2020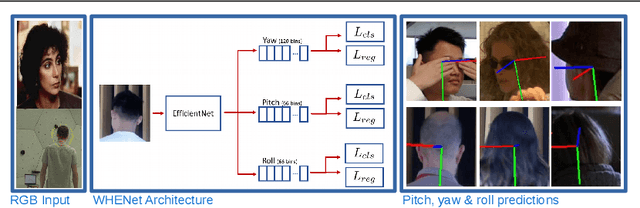
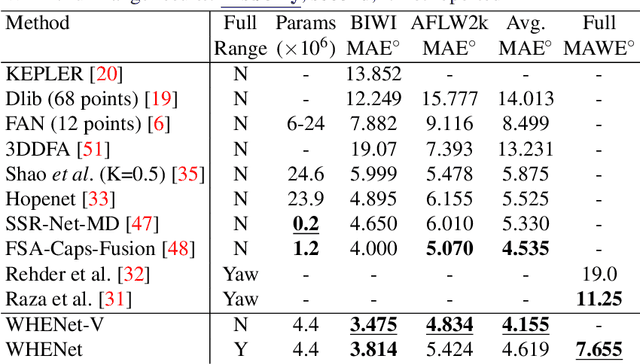
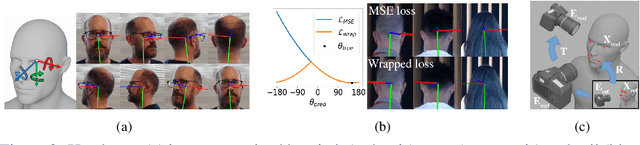
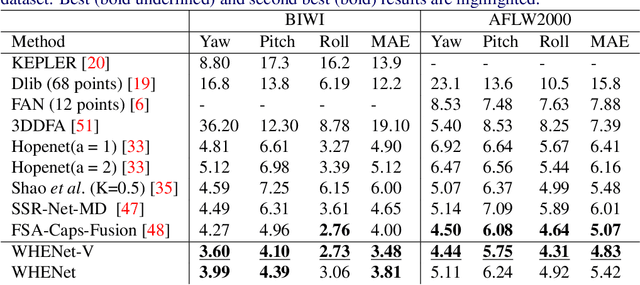
Abstract:We present an end-to-end head-pose estimation network designed to predict Euler angles through the full range head yaws from a single RGB image. Existing methods perform well for frontal views but few target head pose from all viewpoints. This has applications in autonomous driving and retail. Our network builds on multi-loss approaches with changes to loss functions and training strategies adapted to wide range estimation. Additionally, we extract ground truth labelings of anterior views from a current panoptic dataset for the first time. The resulting Wide Headpose Estimation Network (WHENet) is the first fine-grained modern method applicable to the full-range of head yaws (hence wide) yet also meets or beats state-of-the-art methods for frontal head pose estimation. Our network is compact and efficient for mobile devices and applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge