Deep Learning to Predict Late-Onset Breast Cancer Metastasis: the Single Hyperparameter Grid Search (SHGS) Strategy for Meta Tuning Concerning Deep Feed-forward Neural Network
Paper and Code
Aug 28, 2024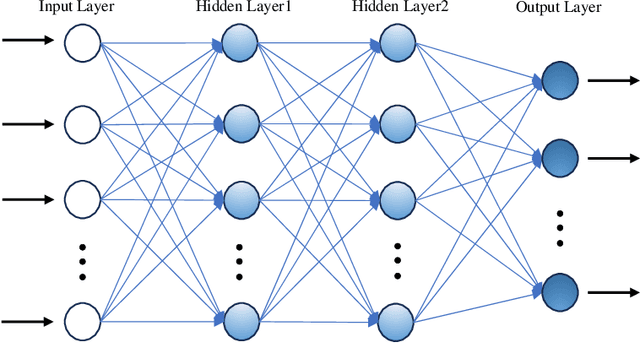

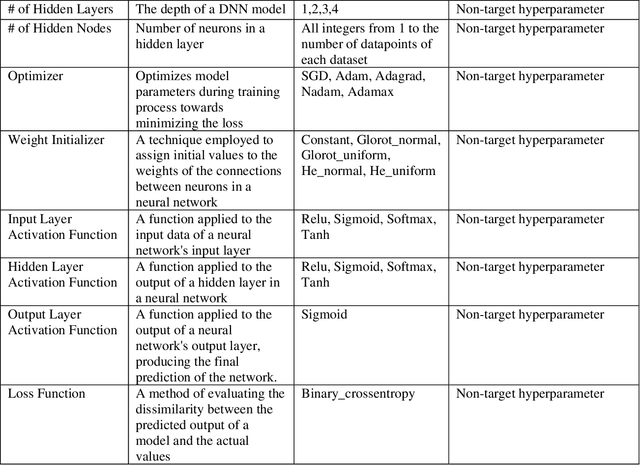
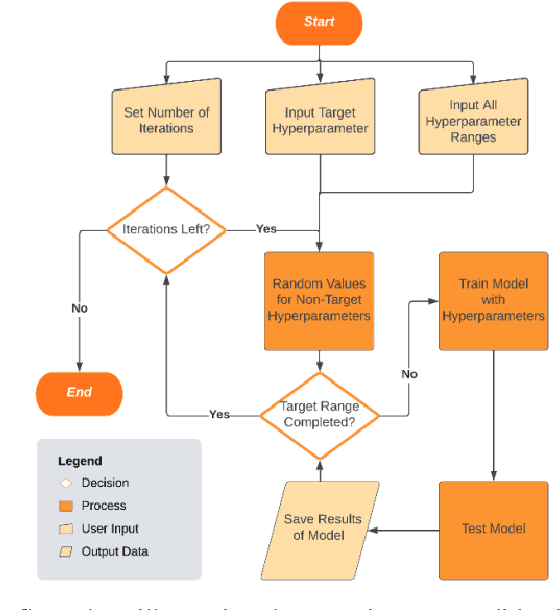
While machine learning has advanced in medicine, its widespread use in clinical applications, especially in predicting breast cancer metastasis, is still limited. We have been dedicated to constructing a DFNN model to predict breast cancer metastasis n years in advance. However, the challenge lies in efficiently identifying optimal hyperparameter values through grid search, given the constraints of time and resources. Issues such as the infinite possibilities for continuous hyperparameters like l1 and l2, as well as the time-consuming and costly process, further complicate the task. To address these challenges, we developed Single Hyperparameter Grid Search (SHGS) strategy, serving as a preselection method before grid search. Our experiments with SHGS applied to DFNN models for breast cancer metastasis prediction focus on analyzing eight target hyperparameters: epochs, batch size, dropout, L1, L2, learning rate, decay, and momentum. We created three figures, each depicting the experiment results obtained from three LSM-I-10-Plus-year datasets. These figures illustrate the relationship between model performance and the target hyperparameter values. For each hyperparameter, we analyzed whether changes in this hyperparameter would affect model performance, examined if there were specific patterns, and explored how to choose values for the particular hyperparameter. Our experimental findings reveal that the optimal value of a hyperparameter is not only dependent on the dataset but is also significantly influenced by the settings of other hyperparameters. Additionally, our experiments suggested some reduced range of values for a target hyperparameter, which may be helpful for low-budget grid search. This approach serves as a prior experience and foundation for subsequent use of grid search to enhance model performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge