Adam Brufsky
Multi-modal AI for comprehensive breast cancer prognostication
Oct 28, 2024Abstract:Treatment selection in breast cancer is guided by molecular subtypes and clinical characteristics. Recurrence risk assessment plays a crucial role in personalizing treatment. Current methods, including genomic assays, have limited accuracy and clinical utility, leading to suboptimal decisions for many patients. We developed a test for breast cancer patient stratification based on digital pathology and clinical characteristics using novel AI methods. Specifically, we utilized a vision transformer-based pan-cancer foundation model trained with self-supervised learning to extract features from digitized H&E-stained slides. These features were integrated with clinical data to form a multi-modal AI test predicting cancer recurrence and death. The test was developed and evaluated using data from a total of 8,161 breast cancer patients across 15 cohorts originating from seven countries. Of these, 3,502 patients from five cohorts were used exclusively for evaluation, while the remaining patients were used for training. Our test accurately predicted our primary endpoint, disease-free interval, in the five external cohorts (C-index: 0.71 [0.68-0.75], HR: 3.63 [3.02-4.37, p<0.01]). In a direct comparison (N=858), the AI test was more accurate than Oncotype DX, the standard-of-care 21-gene assay, with a C-index of 0.67 [0.61-0.74] versus 0.61 [0.49-0.73], respectively. Additionally, the AI test added independent information to Oncotype DX in a multivariate analysis (HR: 3.11 [1.91-5.09, p<0.01)]). The test demonstrated robust accuracy across all major breast cancer subtypes, including TNBC (C-index: 0.71 [0.62-0.81], HR: 3.81 [2.35-6.17, p=0.02]), where no diagnostic tools are currently recommended by clinical guidelines. These results suggest that our AI test can improve accuracy, extend applicability to a wider range of patients, and enhance access to treatment selection tools.
Coalitions of AI-based Methods Predict 15-Year Risks of Breast Cancer Metastasis Using Real-World Clinical Data with AUC up to 0.9
Aug 29, 2024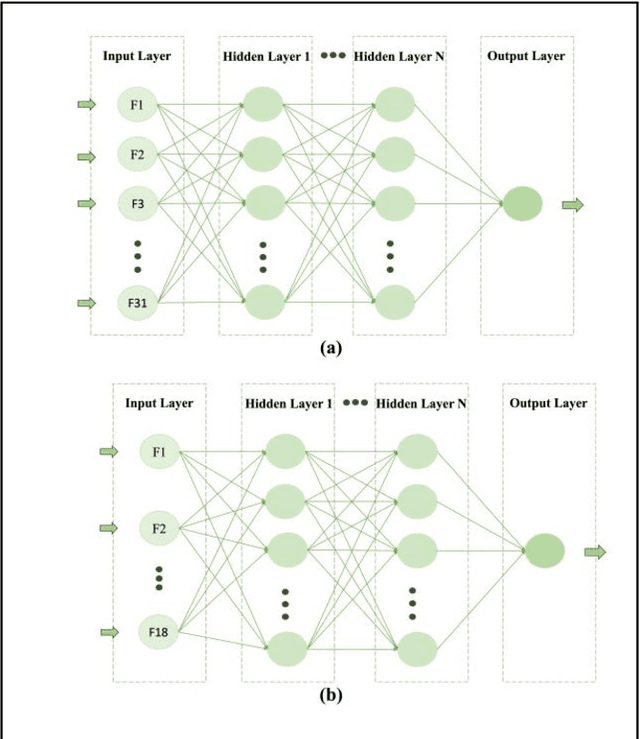
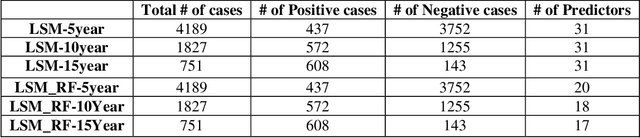
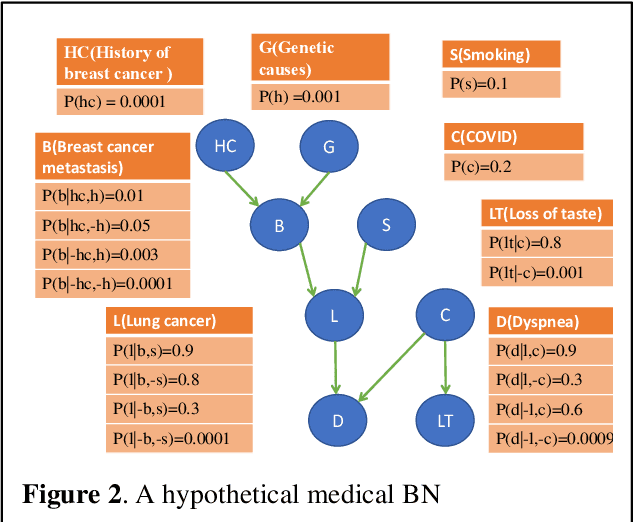
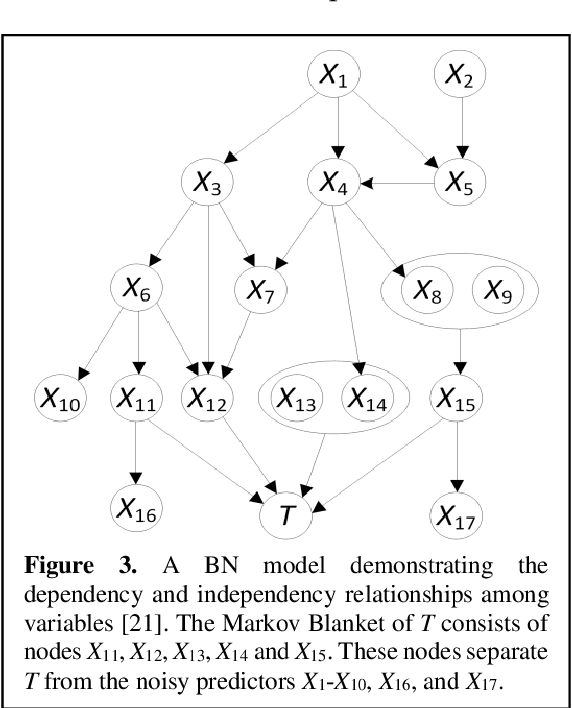
Abstract:Breast cancer is one of the two cancers responsible for the most deaths in women, with about 42,000 deaths each year in the US. That there are over 300,000 breast cancers newly diagnosed each year suggests that only a fraction of the cancers result in mortality. Thus, most of the women undergo seemingly curative treatment for localized cancers, but a significant later succumb to metastatic disease for which current treatments are only temporizing for the vast majority. The current prognostic metrics are of little actionable value for 4 of the 5 women seemingly cured after local treatment, and many women are exposed to morbid and even mortal adjuvant therapies unnecessarily, with these adjuvant therapies reducing metastatic recurrence by only a third. Thus, there is a need for better prognostics to target aggressive treatment at those who are likely to relapse and spare those who were actually cured. While there is a plethora of molecular and tumor-marker assays in use and under-development to detect recurrence early, these are time consuming, expensive and still often un-validated as to actionable prognostic utility. A different approach would use large data techniques to determine clinical and histopathological parameters that would provide accurate prognostics using existing data. Herein, we report on machine learning, together with grid search and Bayesian Networks to develop algorithms that present a AUC of up to 0.9 in ROC analyses, using only extant data. Such algorithms could be rapidly translated to clinical management as they do not require testing beyond routine tumor evaluations.
Deep Learning: a Heuristic Three-stage Mechanism for Grid Searches to Optimize the Future Risk Prediction of Breast Cancer Metastasis Using EHR-based Clinical Data
Aug 15, 2024Abstract:A grid search, at the cost of training and testing a large number of models, is an effective way to optimize the prediction performance of deep learning models. A challenging task concerning grid search is the time management. Without a good time management scheme, a grid search can easily be set off as a mission that will not finish in our lifetime. In this study, we introduce a heuristic three-stage mechanism for managing the running time of low-budget grid searches, and the sweet-spot grid search (SSGS) and randomized grid search (RGS) strategies for improving model prediction performance, in predicting the 5-year, 10-year, and 15-year risk of breast cancer metastasis. We develop deep feedforward neural network (DFNN) models and optimize them through grid searches. We conduct eight cycles of grid searches by applying our three-stage mechanism and SSGS and RGS strategies. We conduct various SHAP analyses including unique ones that interpret the importance of the DFNN-model hyperparameters. Our results show that grid search can greatly improve model prediction. The grid searches we conducted improved the risk prediction of 5-year, 10-year, and 15-year breast cancer metastasis by 18.6%, 16.3%, and 17.3% respectively, over the average performance of all corresponding models we trained using the RGS strategy. We not only demonstrate best model performance but also characterize grid searches from various aspects such as their capabilities of discovering decent models and the unit grid search time. The three-stage mechanism worked effectively. It made our low-budget grid searches feasible and manageable, and in the meantime helped improve model prediction performance. Our SHAP analyses identified both clinical risk factors important for the prediction of future risk of breast cancer metastasis, and DFNN-model hyperparameters important to the prediction of performance scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge