Chuhan Xu
Deep Learning: a Heuristic Three-stage Mechanism for Grid Searches to Optimize the Future Risk Prediction of Breast Cancer Metastasis Using EHR-based Clinical Data
Aug 15, 2024Abstract:A grid search, at the cost of training and testing a large number of models, is an effective way to optimize the prediction performance of deep learning models. A challenging task concerning grid search is the time management. Without a good time management scheme, a grid search can easily be set off as a mission that will not finish in our lifetime. In this study, we introduce a heuristic three-stage mechanism for managing the running time of low-budget grid searches, and the sweet-spot grid search (SSGS) and randomized grid search (RGS) strategies for improving model prediction performance, in predicting the 5-year, 10-year, and 15-year risk of breast cancer metastasis. We develop deep feedforward neural network (DFNN) models and optimize them through grid searches. We conduct eight cycles of grid searches by applying our three-stage mechanism and SSGS and RGS strategies. We conduct various SHAP analyses including unique ones that interpret the importance of the DFNN-model hyperparameters. Our results show that grid search can greatly improve model prediction. The grid searches we conducted improved the risk prediction of 5-year, 10-year, and 15-year breast cancer metastasis by 18.6%, 16.3%, and 17.3% respectively, over the average performance of all corresponding models we trained using the RGS strategy. We not only demonstrate best model performance but also characterize grid searches from various aspects such as their capabilities of discovering decent models and the unit grid search time. The three-stage mechanism worked effectively. It made our low-budget grid searches feasible and manageable, and in the meantime helped improve model prediction performance. Our SHAP analyses identified both clinical risk factors important for the prediction of future risk of breast cancer metastasis, and DFNN-model hyperparameters important to the prediction of performance scores.
Balancing Augmentation with Edge-Utility Filter for Signed GNNs
Oct 25, 2023



Abstract:Signed graph neural networks (SGNNs) has recently drawn more attention as many real-world networks are signed networks containing two types of edges: positive and negative. The existence of negative edges affects the SGNN robustness on two aspects. One is the semantic imbalance as the negative edges are usually hard to obtain though they can provide potentially useful information. The other is the structural unbalance, e.g. unbalanced triangles, an indication of incompatible relationship among nodes. In this paper, we propose a balancing augmentation method to address the above two aspects for SGNNs. Firstly, the utility of each negative edge is measured by calculating its occurrence in unbalanced structures. Secondly, the original signed graph is selectively augmented with the use of (1) an edge perturbation regulator to balance the number of positive and negative edges and to determine the ratio of perturbed edges to original edges and (2) an edge utility filter to remove the negative edges with low utility to make the graph structure more balanced. Finally, a SGNN is trained on the augmented graph which effectively explores the credible relationships. A detailed theoretical analysis is also conducted to prove the effectiveness of each module. Experiments on five real-world datasets in link prediction demonstrate that our method has the advantages of effectiveness and generalization and can significantly improve the performance of SGNN backbones.
iMedBot: A Web-based Intelligent Agent for Healthcare Related Prediction and Deep Learning
Oct 07, 2022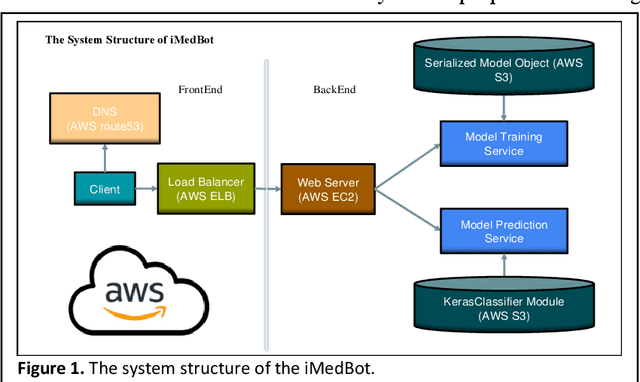
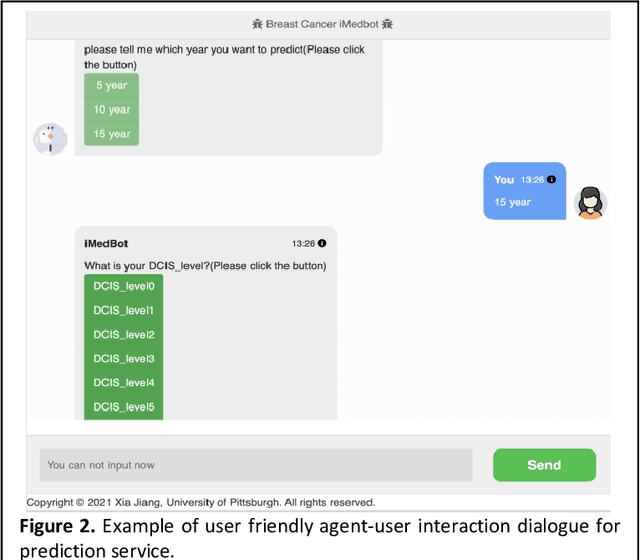
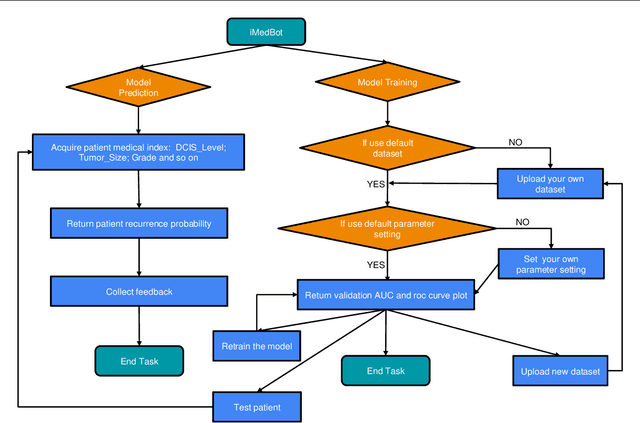
Abstract:Background: Breast cancer is a multifactorial disease, genetic and environmental factors will affect its incidence probability. Breast cancer metastasis is one of the main cause of breast cancer related deaths reported by the American Cancer Society (ACS). Method: the iMedBot is a web application that we developed using the python Flask web framework and deployed on Amazon Web Services. It contains a frontend and a backend. The backend is supported by a python program we developed using the python Keras and scikit-learn packages, which can be used to learn deep feedforward neural network (DFNN) models. Result: the iMedBot can provide two main services: 1. it can predict 5-, 10-, or 15-year breast cancer metastasis based on a set of clinical information provided by a user. The prediction is done by using a set of DFNN models that were pretrained, and 2. It can train DFNN models for a user using user-provided dataset. The model trained will be evaluated using AUC and both the AUC value and the AUC ROC curve will be provided. Conclusion: The iMedBot web application provides a user-friendly interface for user-agent interaction in conducting personalized prediction and model training. It is an initial attempt to convert results of deep learning research into an online tool that may stir further research interests in this direction. Keywords: Deep learning, Breast Cancer, Web application, Model training.
Empirical Study of Overfitting in Deep FNN Prediction Models for Breast Cancer Metastasis
Aug 03, 2022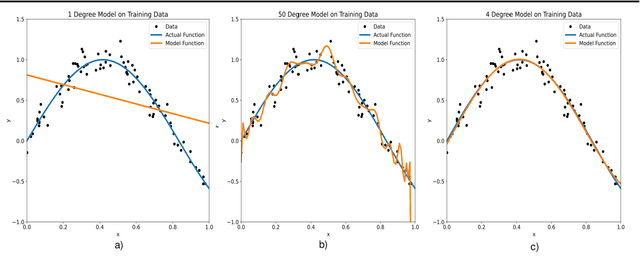
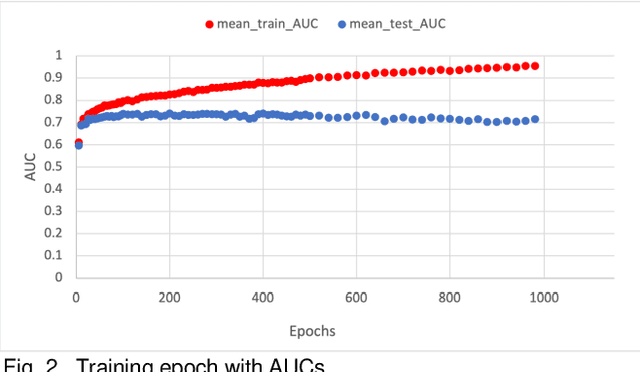
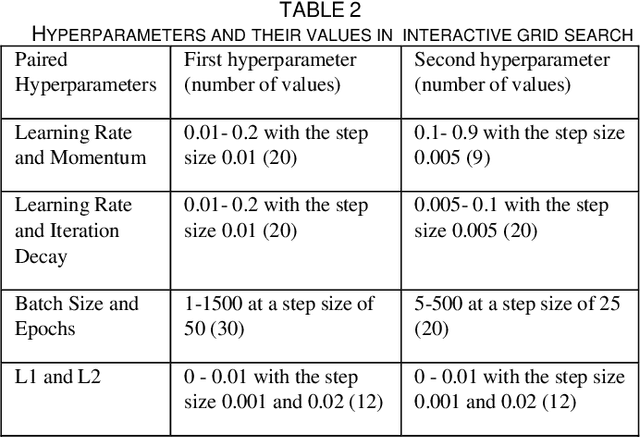
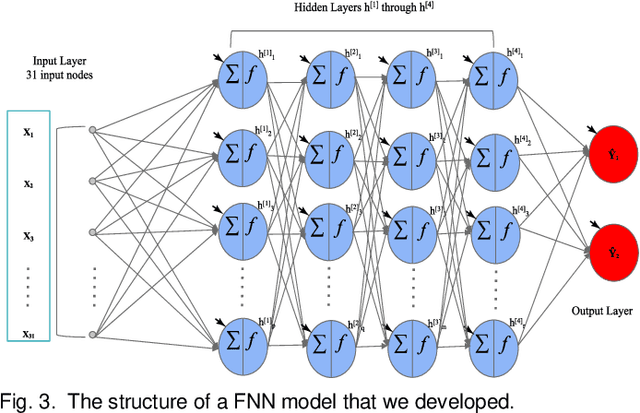
Abstract:Overfitting is defined as the fact that the current model fits a specific data set perfectly, resulting in weakened generalization, and ultimately may affect the accuracy in predicting future data. In this research we used an EHR dataset concerning breast cancer metastasis to study overfitting of deep feedforward Neural Networks (FNNs) prediction models. We included 11 hyperparameters of the deep FNNs models and took an empirical approach to study how each of these hyperparameters was affecting both the prediction performance and overfitting when given a large range of values. We also studied how some of the interesting pairs of hyperparameters were interacting to influence the model performance and overfitting. The 11 hyperparameters we studied include activate function; weight initializer, number of hidden layers, learning rate, momentum, decay, dropout rate, batch size, epochs, L1, and L2. Our results show that most of the single hyperparameters are either negatively or positively corrected with model prediction performance and overfitting. In particular, we found that overfitting overall tends to negatively correlate with learning rate, decay, batch sides, and L2, but tends to positively correlate with momentum, epochs, and L1. According to our results, learning rate, decay, and batch size may have a more significant impact on both overfitting and prediction performance than most of the other hyperparameters, including L1, L2, and dropout rate, which were designed for minimizing overfitting. We also find some interesting interacting pairs of hyperparameters such as learning rate and momentum, learning rate and decay, and batch size and epochs. Keywords: Deep learning, overfitting, prediction, grid search, feedforward neural networks, breast cancer metastasis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge