Yanli Wang
AlignCoder: Aligning Retrieval with Target Intent for Repository-Level Code Completion
Jan 27, 2026Abstract:Repository-level code completion remains a challenging task for existing code large language models (code LLMs) due to their limited understanding of repository-specific context and domain knowledge. While retrieval-augmented generation (RAG) approaches have shown promise by retrieving relevant code snippets as cross-file context, they suffer from two fundamental problems: misalignment between the query and the target code in the retrieval process, and the inability of existing retrieval methods to effectively utilize the inference information. To address these challenges, we propose AlignCoder, a repository-level code completion framework that introduces a query enhancement mechanism and a reinforcement learning based retriever training method. Our approach generates multiple candidate completions to construct an enhanced query that bridges the semantic gap between the initial query and the target code. Additionally, we employ reinforcement learning to train an AlignRetriever that learns to leverage inference information in the enhanced query for more accurate retrieval. We evaluate AlignCoder on two widely-used benchmarks (CrossCodeEval and RepoEval) across five backbone code LLMs, demonstrating an 18.1% improvement in EM score compared to baselines on the CrossCodeEval benchmark. The results show that our framework achieves superior performance and exhibits high generalizability across various code LLMs and programming languages.
Towards an Understanding of Context Utilization in Code Intelligence
Apr 11, 2025Abstract:Code intelligence is an emerging domain in software engineering, aiming to improve the effectiveness and efficiency of various code-related tasks. Recent research suggests that incorporating contextual information beyond the basic original task inputs (i.e., source code) can substantially enhance model performance. Such contextual signals may be obtained directly or indirectly from sources such as API documentation or intermediate representations like abstract syntax trees can significantly improve the effectiveness of code intelligence. Despite growing academic interest, there is a lack of systematic analysis of context in code intelligence. To address this gap, we conduct an extensive literature review of 146 relevant studies published between September 2007 and August 2024. Our investigation yields four main contributions. (1) A quantitative analysis of the research landscape, including publication trends, venues, and the explored domains; (2) A novel taxonomy of context types used in code intelligence; (3) A task-oriented analysis investigating context integration strategies across diverse code intelligence tasks; (4) A critical evaluation of evaluation methodologies for context-aware methods. Based on these findings, we identify fundamental challenges in context utilization in current code intelligence systems and propose a research roadmap that outlines key opportunities for future research.
RepoTransBench: A Real-World Benchmark for Repository-Level Code Translation
Dec 23, 2024



Abstract:Repository-level code translation refers to translating an entire code repository from one programming language to another while preserving the functionality of the source repository. Many benchmarks have been proposed to evaluate the performance of such code translators. However, previous benchmarks mostly provide fine-grained samples, focusing at either code snippet, function, or file-level code translation. Such benchmarks do not accurately reflect real-world demands, where entire repositories often need to be translated, involving longer code length and more complex functionalities. To address this gap, we propose a new benchmark, named RepoTransBench, which is a real-world repository-level code translation benchmark with an automatically executable test suite. We conduct experiments on RepoTransBench to evaluate the translation performance of 11 advanced LLMs. We find that the Success@1 score (test success in one attempt) of the best-performing LLM is only 7.33%. To further explore the potential of LLMs for repository-level code translation, we provide LLMs with error-related feedback to perform iterative debugging and observe an average 7.09% improvement on Success@1. However, even with this improvement, the Success@1 score of the best-performing LLM is only 21%, which may not meet the need for reliable automatic repository-level code translation. Finally, we conduct a detailed error analysis and highlight current LLMs' deficiencies in repository-level code translation, which could provide a reference for further improvements.
Diffusion Models in Bioinformatics: A New Wave of Deep Learning Revolution in Action
Feb 13, 2023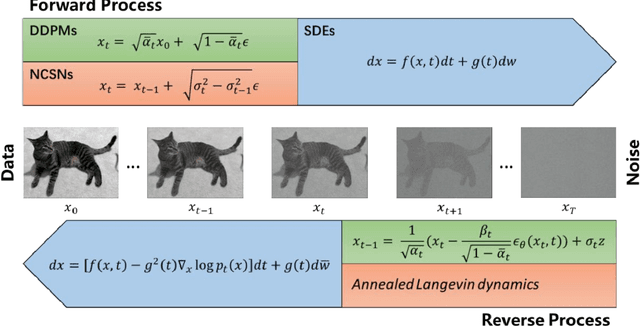
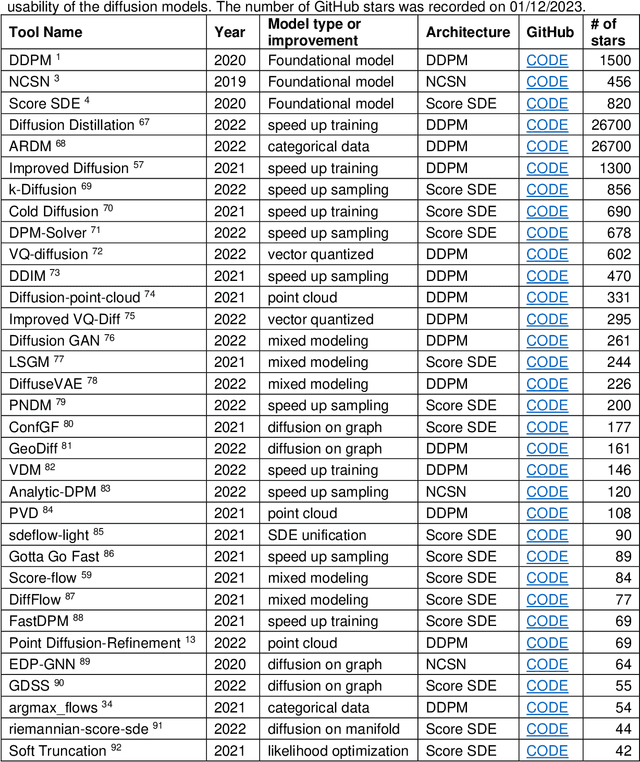
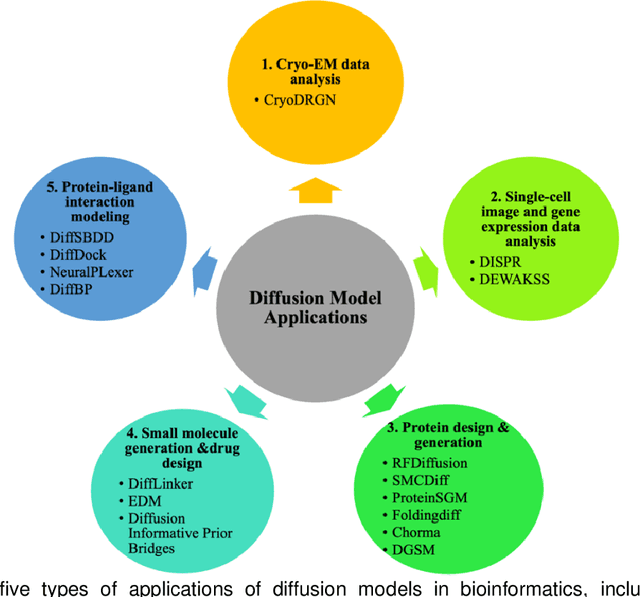
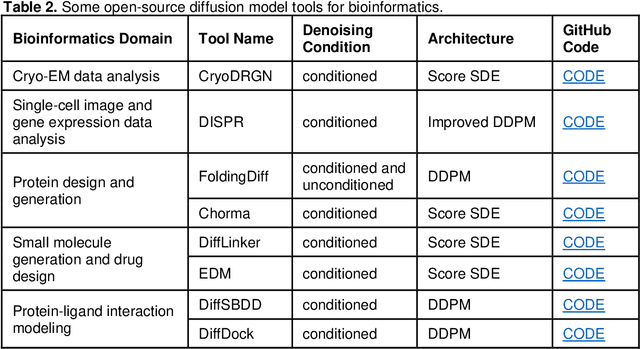
Abstract:Denoising diffusion models have emerged as one of the most powerful generative models in recent years. They have achieved remarkable success in many fields, such as computer vision, natural language processing (NLP), and bioinformatics. Although there are a few excellent reviews on diffusion models and their applications in computer vision and NLP, there is a lack of an overview of their applications in bioinformatics. This review aims to provide a rather thorough overview of the applications of diffusion models in bioinformatics to aid their further development in bioinformatics and computational biology. We start with an introduction of the key concepts and theoretical foundations of three cornerstone diffusion modeling frameworks (denoising diffusion probabilistic models, noise-conditioned scoring networks, and stochastic differential equations), followed by a comprehensive description of diffusion models employed in the different domains of bioinformatics, including cryo-EM data enhancement, single-cell data analysis, protein design and generation, drug and small molecule design, and protein-ligand interaction. The review is concluded with a summary of the potential new development and applications of diffusion models in bioinformatics.
Heat Kernel Smoothing in Irregular Image Domains
Oct 21, 2017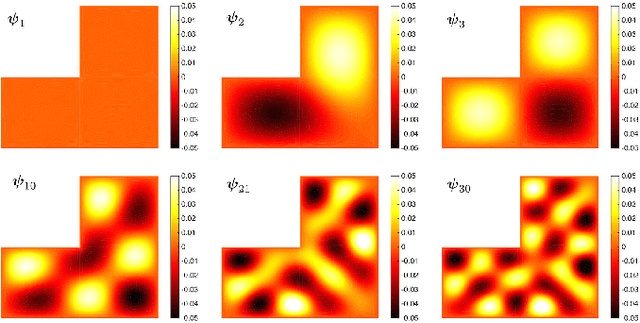
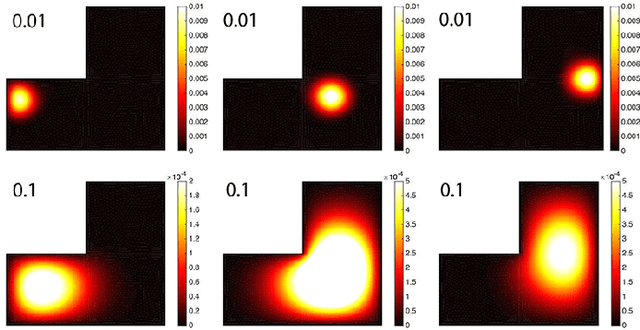
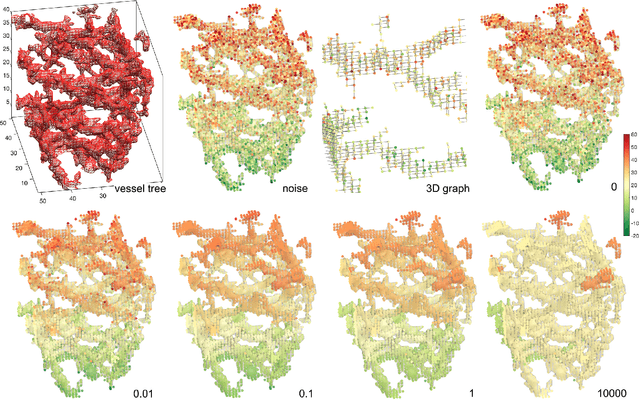
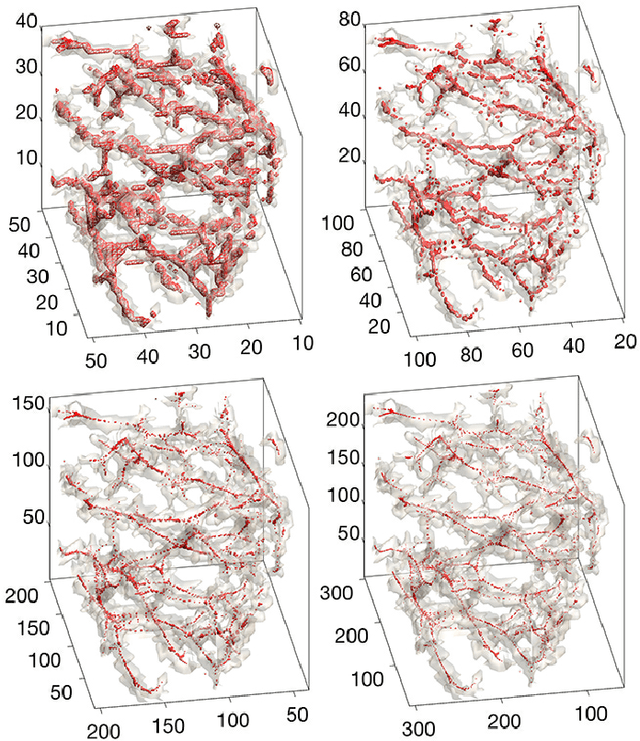
Abstract:We present the discrete version of heat kernel smoothing on graph data structure. The method is used to smooth data in an irregularly shaped domains in 3D images. New statistical properties are derived. As an application, we show how to filter out data in the lung blood vessel trees obtained from computed tomography. The method can be further used in representing the complex vessel trees parametrically and extracting the skeleton representation of the trees.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge