Xiang Sun
FusionMAE: large-scale pretrained model to optimize and simplify diagnostic and control of fusion plasma
Sep 16, 2025Abstract:In magnetically confined fusion device, the complex, multiscale, and nonlinear dynamics of plasmas necessitate the integration of extensive diagnostic systems to effectively monitor and control plasma behaviour. The complexity and uncertainty arising from these extensive systems and their tangled interrelations has long posed a significant obstacle to the acceleration of fusion energy development. In this work, a large-scale model, fusion masked auto-encoder (FusionMAE) is pre-trained to compress the information from 88 diagnostic signals into a concrete embedding, to provide a unified interface between diagnostic systems and control actuators. Two mechanisms are proposed to ensure a meaningful embedding: compression-reduction and missing-signal reconstruction. Upon completion of pre-training, the model acquires the capability for 'virtual backup diagnosis', enabling the inference of missing diagnostic data with 96.7% reliability. Furthermore, the model demonstrates three emergent capabilities: automatic data analysis, universal control-diagnosis interface, and enhancement of control performance on multiple tasks. This work pioneers large-scale AI model integration in fusion energy, demonstrating how pre-trained embeddings can simplify the system interface, reducing necessary diagnostic systems and optimize operation performance for future fusion reactors.
LONGER: Scaling Up Long Sequence Modeling in Industrial Recommenders
May 07, 2025

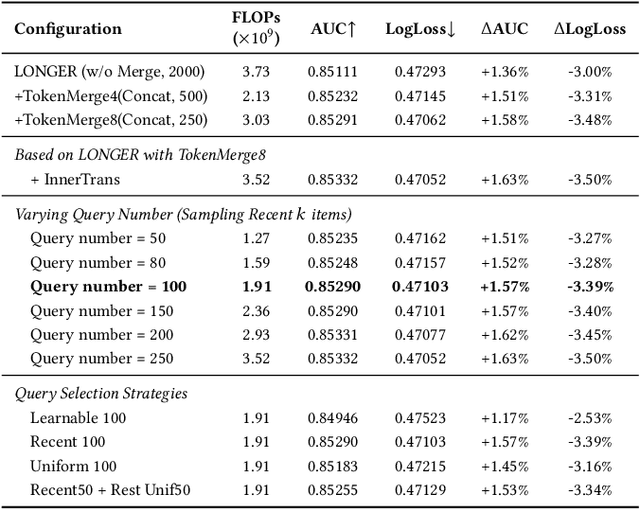
Abstract:Modeling ultra-long user behavior sequences is critical for capturing both long- and short-term preferences in industrial recommender systems. Existing solutions typically rely on two-stage retrieval or indirect modeling paradigms, incuring upstream-downstream inconsistency and computational inefficiency. In this paper, we present LONGER, a Long-sequence Optimized traNsformer for GPU-Efficient Recommenders. LONGER incorporates (i) a global token mechanism for stabilizing attention over long contexts, (ii) a token merge module with lightweight InnerTransformers and hybrid attention strategy to reduce quadratic complexity, and (iii) a series of engineering optimizations, including training with mixed-precision and activation recomputation, KV cache serving, and the fully synchronous model training and serving framework for unified GPU-based dense and sparse parameter updates. LONGER consistently outperforms strong baselines in both offline metrics and online A/B testing in both advertising and e-commerce services at ByteDance, validating its consistent effectiveness and industrial-level scaling laws. Currently, LONGER has been fully deployed at more than 10 influential scenarios at ByteDance, serving billion users.
Sky of Unlearning (SoUL): Rewiring Federated Machine Unlearning via Selective Pruning
Apr 02, 2025



Abstract:The Internet of Drones (IoD), where drones collaborate in data collection and analysis, has become essential for applications such as surveillance and environmental monitoring. Federated learning (FL) enables drones to train machine learning models in a decentralized manner while preserving data privacy. However, FL in IoD networks is susceptible to attacks like data poisoning and model inversion. Federated unlearning (FU) mitigates these risks by eliminating adversarial data contributions, preventing their influence on the model. This paper proposes sky of unlearning (SoUL), a federated unlearning framework that efficiently removes the influence of unlearned data while maintaining model performance. A selective pruning algorithm is designed to identify and remove neurons influential in unlearning but minimally impact the overall performance of the model. Simulations demonstrate that SoUL outperforms existing unlearning methods, achieves accuracy comparable to full retraining, and reduces computation and communication overhead, making it a scalable and efficient solution for resource-constrained IoD networks.
GraphGANFed: A Federated Generative Framework for Graph-Structured Molecules Towards Efficient Drug Discovery
Apr 11, 2023



Abstract:Recent advances in deep learning have accelerated its use in various applications, such as cellular image analysis and molecular discovery. In molecular discovery, a generative adversarial network (GAN), which comprises a discriminator to distinguish generated molecules from existing molecules and a generator to generate new molecules, is one of the premier technologies due to its ability to learn from a large molecular data set efficiently and generate novel molecules that preserve similar properties. However, different pharmaceutical companies may be unwilling or unable to share their local data sets due to the geo-distributed and sensitive nature of molecular data sets, making it impossible to train GANs in a centralized manner. In this paper, we propose a Graph convolutional network in Generative Adversarial Networks via Federated learning (GraphGANFed) framework, which integrates graph convolutional neural Network (GCN), GAN, and federated learning (FL) as a whole system to generate novel molecules without sharing local data sets. In GraphGANFed, the discriminator is implemented as a GCN to better capture features from molecules represented as molecular graphs, and FL is used to train both the discriminator and generator in a distributive manner to preserve data privacy. Extensive simulations are conducted based on the three bench-mark data sets to demonstrate the feasibility and effectiveness of GraphGANFed. The molecules generated by GraphGANFed can achieve high novelty (=100) and diversity (> 0.9). The simulation results also indicate that 1) a lower complexity discriminator model can better avoid mode collapse for a smaller data set, 2) there is a tradeoff among different evaluation metrics, and 3) having the right dropout ratio of the generator and discriminator can avoid mode collapse.
Latency Aware Semi-synchronous Client Selection and Model Aggregation for Wireless Federated Learning
Oct 19, 2022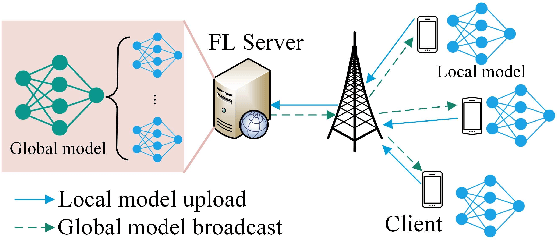

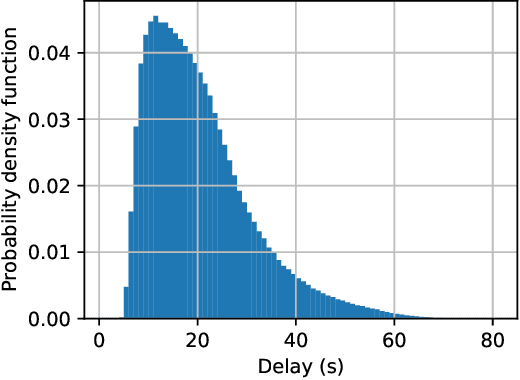
Abstract:Federated learning (FL) is a collaborative machine learning framework that requires different clients (e.g., Internet of Things devices) to participate in the machine learning model training process by training and uploading their local models to an FL server in each global iteration. Upon receiving the local models from all the clients, the FL server generates a global model by aggregating the received local models. This traditional FL process may suffer from the straggler problem in heterogeneous client settings, where the FL server has to wait for slow clients to upload their local models in each global iteration, thus increasing the overall training time. One of the solutions is to set up a deadline and only the clients that can upload their local models before the deadline would be selected in the FL process. This solution may lead to a slow convergence rate and global model overfitting issues due to the limited client selection. In this paper, we propose the Latency awarE Semi-synchronous client Selection and mOdel aggregation for federated learNing (LESSON) method that allows all the clients to participate in the whole FL process but with different frequencies. That is, faster clients would be scheduled to upload their models more frequently than slow clients, thus resolving the straggler problem and accelerating the convergence speed, while avoiding model overfitting. Also, LESSON is capable of adjusting the tradeoff between the model accuracy and convergence rate by varying the deadline. Extensive simulations have been conducted to compare the performance of LESSON with the other two baseline methods, i.e., FedAvg and FedCS. The simulation results demonstrate that LESSON achieves faster convergence speed than FedAvg and FedCS, and higher model accuracy than FedCS.
Backhaul-Aware Drone Base Station Placement and Resource Management for FSO based Drone Assisted Mobile Networks
Dec 24, 2021



Abstract:In drone assisted mobile networks, drones mounted small cell base stations (DBSs) are responsively and flexibly deployed over any Places of Interest (PoI), such as sporadic hotspots and disaster-struck areas, where the existing mobile network infrastructure is unable to provide wireless coverage. Here, a DBS is a relay node to relay traffic between a nearby macro base station (MBS) and the users. In addition, Free-space optics (FSO) is applied as the backhauling solution to significantly increase the capacity of the backhaul link between an MBS and a DBS in a drone assisted mobile network. Most of the existing DBS placement solutions assume the FSO based backhaul link provides sufficient link capacity, which may not be true, especially when a DBS is placed far away from an MBS (e.g., > 10 km in disaster-struck areas) or in a bad weather condition. In this paper, we formulate a problem to jointly optimize bandwidth allocation and DBS placement by considering the FSO based backhaul link capacity constraint. A Backhaul awaRe bandwidth allOcAtion and DBS placement (BROAD) algorithm is designed to efficiently solve the problem, and the performance of the algorithm is demonstrated via extensive simulations.
Computing Cliques and Cavities in Networks
Jan 03, 2021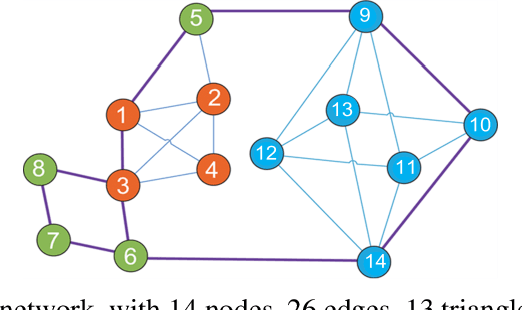

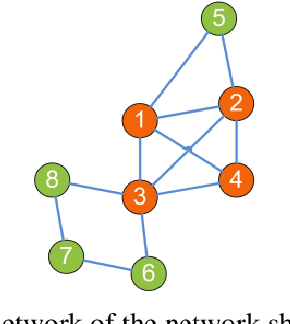
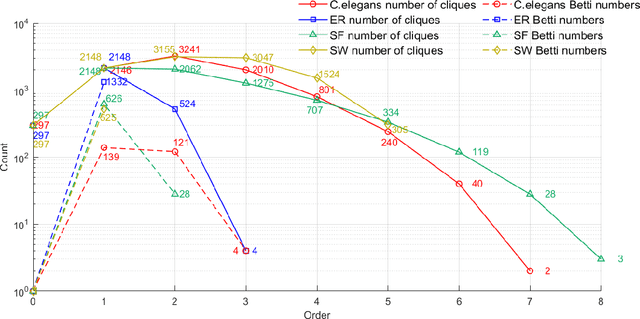
Abstract:Complex networks have complete subgraphs such as nodes, edges, triangles, etc., referred to as cliques of different orders. Notably, cavities consisting of higher-order cliques have been found playing an important role in brain functions. Since searching for the maximum clique in a large network is an NP-complete problem, we propose using k-core decomposition to determine the computability of a given network subject to limited computing resources. For a computable network, we design a search algorithm for finding cliques of different orders, which also provides the Euler characteristic number. Then, we compute the Betti number by using the ranks of the boundary matrices of adjacent cliques. Furthermore, we design an optimized algorithm for finding cavities of different orders. Finally, we apply the algorithm to the neuronal network of C. elegans in one dataset, and find its all cliques and some cavities of different orders therein, providing a basis for further mathematical analysis and computation of the structure and function of the C. elegans neuronal network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge