Wongi Park
Veta-GS: View-dependent deformable 3D Gaussian Splatting for thermal infrared Novel-view Synthesis
May 25, 2025



Abstract:Recently, 3D Gaussian Splatting (3D-GS) based on Thermal Infrared (TIR) imaging has gained attention in novel-view synthesis, showing real-time rendering. However, novel-view synthesis with thermal infrared images suffers from transmission effects, emissivity, and low resolution, leading to floaters and blur effects in rendered images. To address these problems, we introduce Veta-GS, which leverages a view-dependent deformation field and a Thermal Feature Extractor (TFE) to precisely capture subtle thermal variations and maintain robustness. Specifically, we design view-dependent deformation field that leverages camera position and viewing direction, which capture thermal variations. Furthermore, we introduce the Thermal Feature Extractor (TFE) and MonoSSIM loss, which consider appearance, edge, and frequency to maintain robustness. Extensive experiments on the TI-NSD benchmark show that our method achieves better performance over existing methods.
ForestSplats: Deformable transient field for Gaussian Splatting in the Wild
Mar 08, 2025Abstract:Recently, 3D Gaussian Splatting (3D-GS) has emerged, showing real-time rendering speeds and high-quality results in static scenes. Although 3D-GS shows effectiveness in static scenes, their performance significantly degrades in real-world environments due to transient objects, lighting variations, and diverse levels of occlusion. To tackle this, existing methods estimate occluders or transient elements by leveraging pre-trained models or integrating additional transient field pipelines. However, these methods still suffer from two defects: 1) Using semantic features from the Vision Foundation model (VFM) causes additional computational costs. 2) The transient field requires significant memory to handle transient elements with per-view Gaussians and struggles to define clear boundaries for occluders, solely relying on photometric errors. To address these problems, we propose ForestSplats, a novel approach that leverages the deformable transient field and a superpixel-aware mask to efficiently represent transient elements in the 2D scene across unconstrained image collections and effectively decompose static scenes from transient distractors without VFM. We designed the transient field to be deformable, capturing per-view transient elements. Furthermore, we introduce a superpixel-aware mask that clearly defines the boundaries of occluders by considering photometric errors and superpixels. Additionally, we propose uncertainty-aware densification to avoid generating Gaussians within the boundaries of occluders during densification. Through extensive experiments across several benchmark datasets, we demonstrate that ForestSplats outperforms existing methods without VFM and shows significant memory efficiency in representing transient elements.
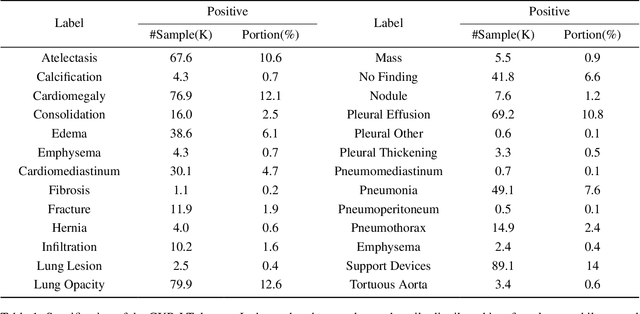
Towards long-tailed, multi-label disease classification from chest X-ray: Overview of the CXR-LT challenge
Oct 24, 2023



Abstract:Many real-world image recognition problems, such as diagnostic medical imaging exams, are "long-tailed" $\unicode{x2013}$ there are a few common findings followed by many more relatively rare conditions. In chest radiography, diagnosis is both a long-tailed and multi-label problem, as patients often present with multiple findings simultaneously. While researchers have begun to study the problem of long-tailed learning in medical image recognition, few have studied the interaction of label imbalance and label co-occurrence posed by long-tailed, multi-label disease classification. To engage with the research community on this emerging topic, we conducted an open challenge, CXR-LT, on long-tailed, multi-label thorax disease classification from chest X-rays (CXRs). We publicly release a large-scale benchmark dataset of over 350,000 CXRs, each labeled with at least one of 26 clinical findings following a long-tailed distribution. We synthesize common themes of top-performing solutions, providing practical recommendations for long-tailed, multi-label medical image classification. Finally, we use these insights to propose a path forward involving vision-language foundation models for few- and zero-shot disease classification.
Robust Asymmetric Loss for Multi-Label Long-Tailed Learning
Aug 10, 2023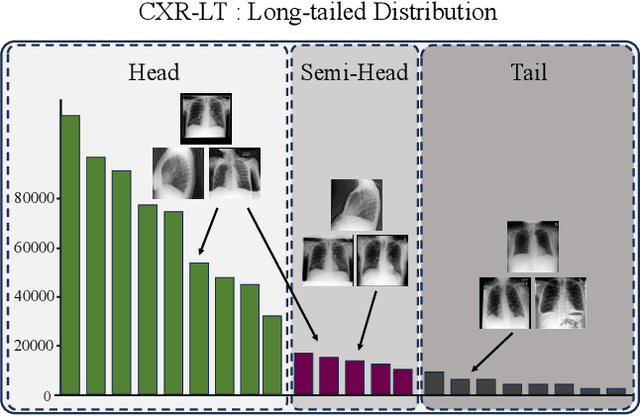
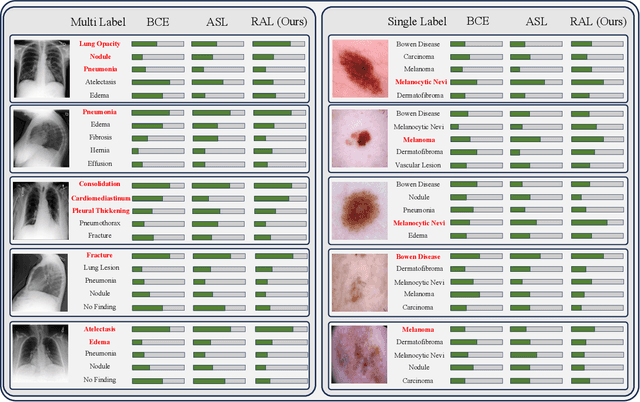
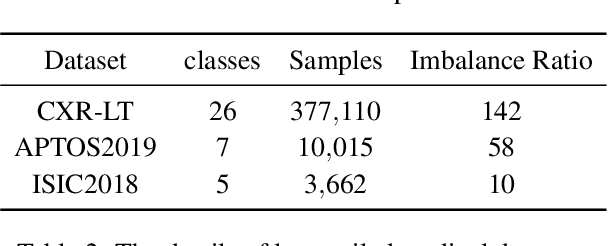
Abstract:In real medical data, training samples typically show long-tailed distributions with multiple labels. Class distribution of the medical data has a long-tailed shape, in which the incidence of different diseases is quite varied, and at the same time, it is not unusual for images taken from symptomatic patients to be multi-label diseases. Therefore, in this paper, we concurrently address these two issues by putting forth a robust asymmetric loss on the polynomial function. Since our loss tackles both long-tailed and multi-label classification problems simultaneously, it leads to a complex design of the loss function with a large number of hyper-parameters. Although a model can be highly fine-tuned due to a large number of hyper-parameters, it is difficult to optimize all hyper-parameters at the same time, and there might be a risk of overfitting a model. Therefore, we regularize the loss function using the Hill loss approach, which is beneficial to be less sensitive against the numerous hyper-parameters so that it reduces the risk of overfitting the model. For this reason, the proposed loss is a generic method that can be applied to most medical image classification tasks and does not make the training process more time-consuming. We demonstrate that the proposed robust asymmetric loss performs favorably against the long-tailed with multi-label medical image classification in addition to the various long-tailed single-label datasets. Notably, our method achieves Top-5 results on the CXR-LT dataset of the ICCV CVAMD 2023 competition. We opensource our implementation of the robust asymmetric loss in the public repository: https://github.com/kalelpark/RAL.
Fine-Grained Self-Supervised Learning with Jigsaw Puzzles for Medical Image Classification
Aug 10, 2023
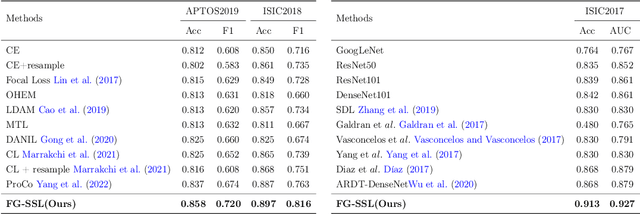
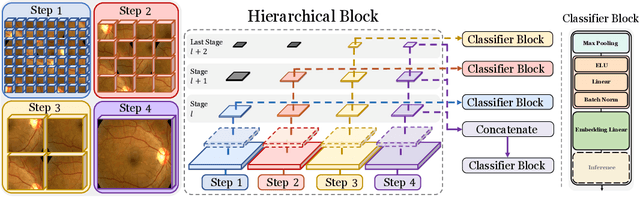
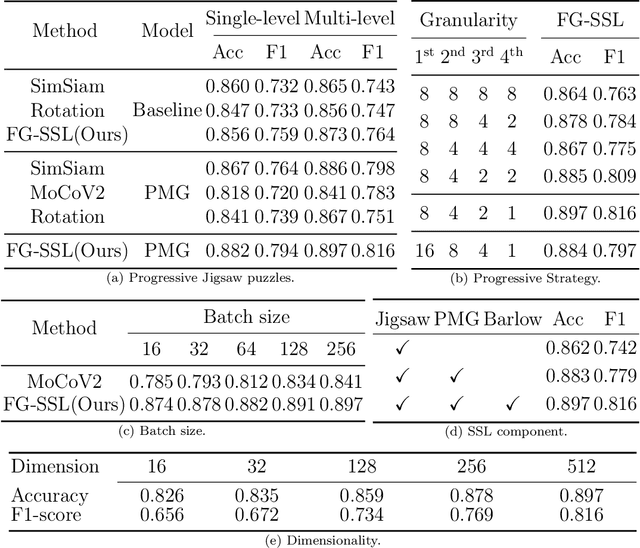
Abstract:Classifying fine-grained lesions is challenging due to minor and subtle differences in medical images. This is because learning features of fine-grained lesions with highly minor differences is very difficult in training deep neural networks. Therefore, in this paper, we introduce Fine-Grained Self-Supervised Learning(FG-SSL) method for classifying subtle lesions in medical images. The proposed method progressively learns the model through hierarchical block such that the cross-correlation between the fine-grained Jigsaw puzzle and regularized original images is close to the identity matrix. We also apply hierarchical block for progressive fine-grained learning, which extracts different information in each step, to supervised learning for discovering subtle differences. Our method does not require an asymmetric model, nor does a negative sampling strategy, and is not sensitive to batch size. We evaluate the proposed fine-grained self-supervised learning method on comprehensive experiments using various medical image recognition datasets. In our experiments, the proposed method performs favorably compared to existing state-of-the-art approaches on the widely-used ISIC2018, APTOS2019, and ISIC2017 datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge