Wenbin Liao
TopoLogic: An Interpretable Pipeline for Lane Topology Reasoning on Driving Scenes
May 23, 2024



Abstract:As an emerging task that integrates perception and reasoning, topology reasoning in autonomous driving scenes has recently garnered widespread attention. However, existing work often emphasizes "perception over reasoning": they typically boost reasoning performance by enhancing the perception of lanes and directly adopt MLP to learn lane topology from lane query. This paradigm overlooks the geometric features intrinsic to the lanes themselves and are prone to being influenced by inherent endpoint shifts in lane detection. To tackle this issue, we propose an interpretable method for lane topology reasoning based on lane geometric distance and lane query similarity, named TopoLogic. This method mitigates the impact of endpoint shifts in geometric space, and introduces explicit similarity calculation in semantic space as a complement. By integrating results from both spaces, our methods provides more comprehensive information for lane topology. Ultimately, our approach significantly outperforms the existing state-of-the-art methods on the mainstream benchmark OpenLane-V2 (23.9 v.s. 10.9 in TOP$_{ll}$ and 44.1 v.s. 39.8 in OLS on subset_A. Additionally, our proposed geometric distance topology reasoning method can be incorporated into well-trained models without re-training, significantly boost the performance of lane topology reasoning. The code is released at https://github.com/Franpin/TopoLogic.
Taxonomy and evolution predicting using deep learning in images
Jun 28, 2022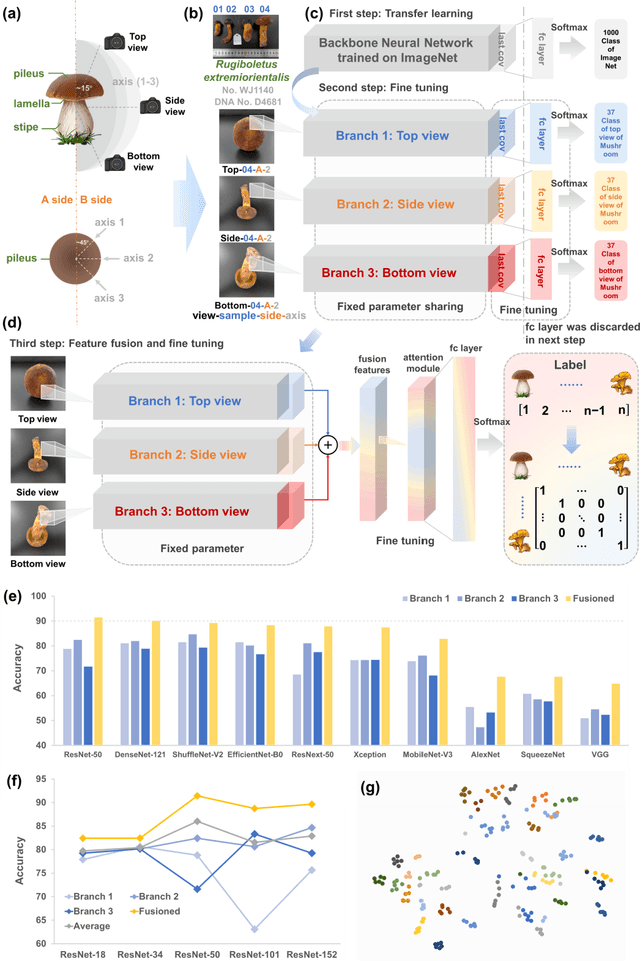
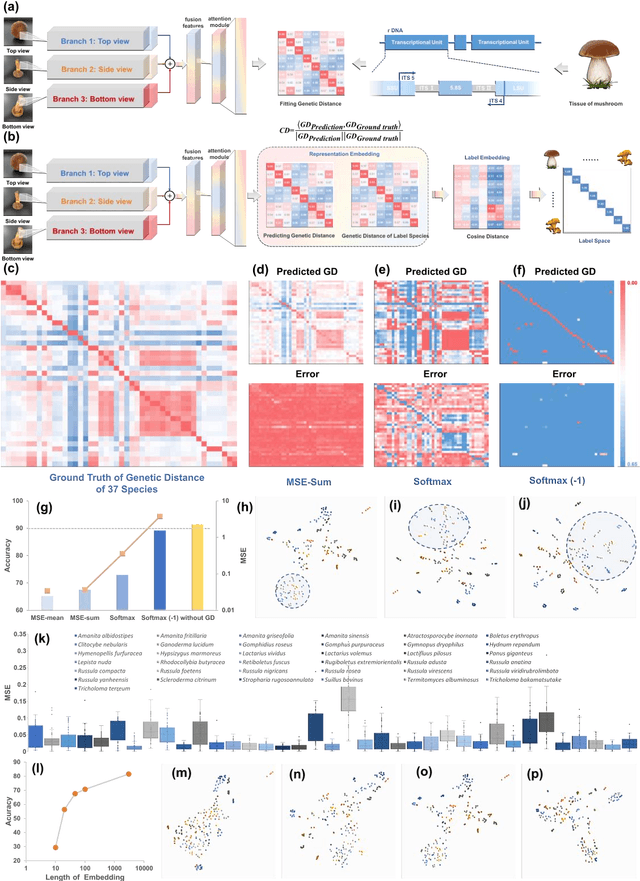
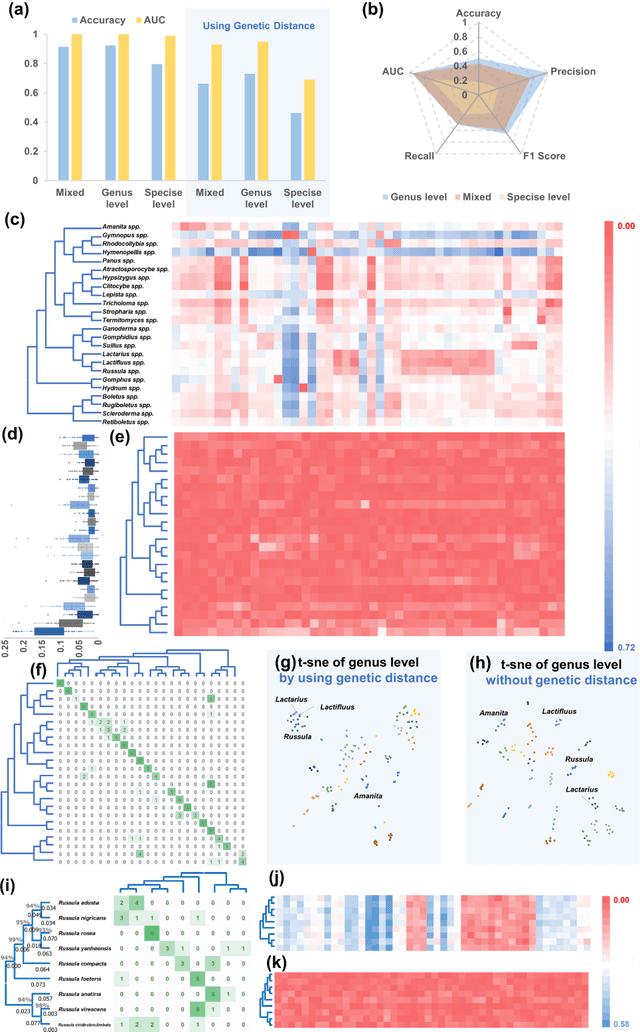
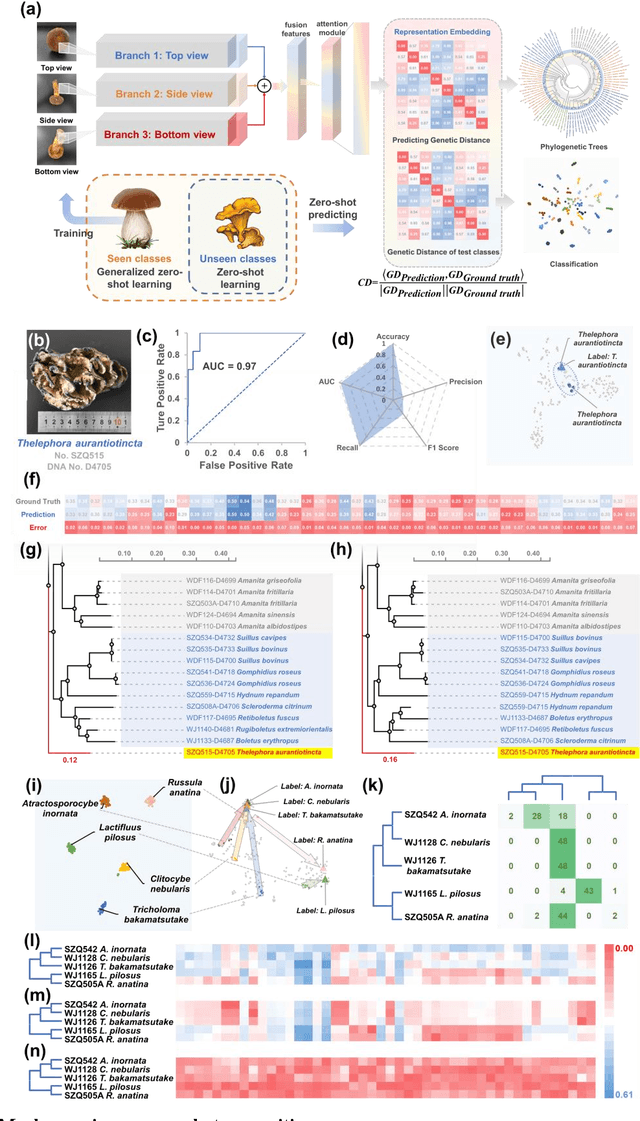
Abstract:Molecular and morphological characters, as important parts of biological taxonomy, are contradictory but need to be integrated. Organism's image recognition and bioinformatics are emerging and hot problems nowadays but with a gap between them. In this work, a multi-branching recognition framework mediated by genetic information bridges this barrier, which establishes the link between macro-morphology and micro-molecular information of mushrooms. The novel multi-perspective structure is proposed to fuse the feature images from three branching models, which significantly improves the accuracy of recognition by about 10% and up to more than 90%. Further, genetic information is implemented to the mushroom image recognition task by using genetic distance embeddings as the representation space for predicting image distance and species identification. Semantic overfitting of traditional classification tasks and the granularity of fine-grained image recognition are also discussed in depth for the first time. The generalizability of the model was investigated in fine-grained scenarios using zero-shot learning tasks, which could predict the taxonomic and evolutionary information of unseen samples. We presented the first method to map images to DNA, namely used an encoder mapping image to genetic distances, and then decoded DNA through a pre-trained decoder, where the total test accuracy on 37 species for DNA prediction is 87.45%. This study creates a novel recognition framework by systematically studying the mushroom image recognition problem, bridging the gap between macroscopic biological information and microscopic molecular information, which will provide a new reference for intelligent biometrics in the future.
Mushroom image recognition and distance generation based on attention-mechanism model and genetic information
Jun 27, 2022
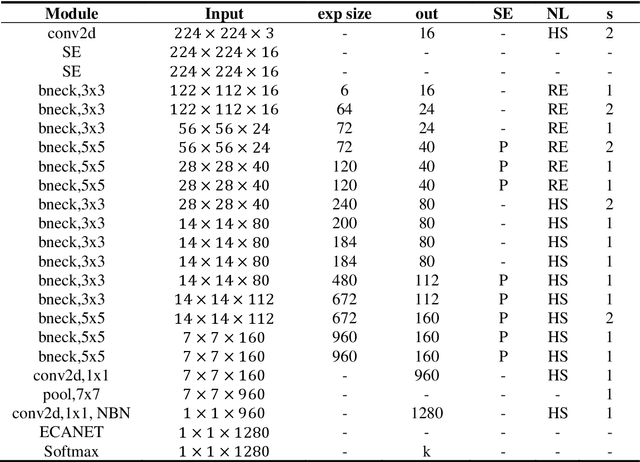

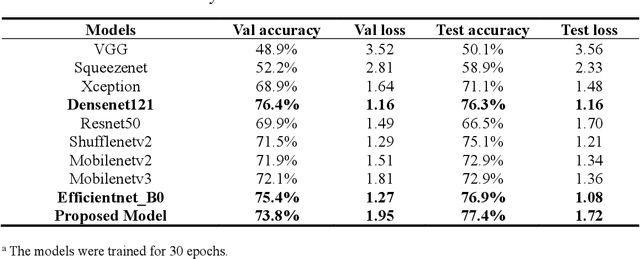
Abstract:The species identification of Macrofungi, i.e. mushrooms, has always been a challenging task. There are still a large number of poisonous mushrooms that have not been found, which poses a risk to people's life. However, the traditional identification method requires a large number of experts with knowledge in the field of taxonomy for manual identification, it is not only inefficient but also consumes a lot of manpower and capital costs. In this paper, we propose a new model based on attention-mechanism, MushroomNet, which applies the lightweight network MobileNetV3 as the backbone model, combined with the attention structure proposed by us, and has achieved excellent performance in the mushroom recognition task. On the public dataset, the test accuracy of the MushroomNet model has reached 83.9%, and on the local dataset, the test accuracy has reached 77.4%. The proposed attention mechanisms well focused attention on the bodies of mushroom image for mixed channel attention and the attention heat maps visualized by Grad-CAM. Further, in this study, genetic distance was added to the mushroom image recognition task, the genetic distance was used as the representation space, and the genetic distance between each pair of mushroom species in the dataset was used as the embedding of the genetic distance representation space, so as to predict the image distance and species. identify. We found that using the MES activation function can predict the genetic distance of mushrooms very well, but the accuracy is lower than that of SoftMax. The proposed MushroomNet was demonstrated it shows great potential for automatic and online mushroom image and the proposed automatic procedure would assist and be a reference to traditional mushroom classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge