Taxonomy and evolution predicting using deep learning in images
Paper and Code
Jun 28, 2022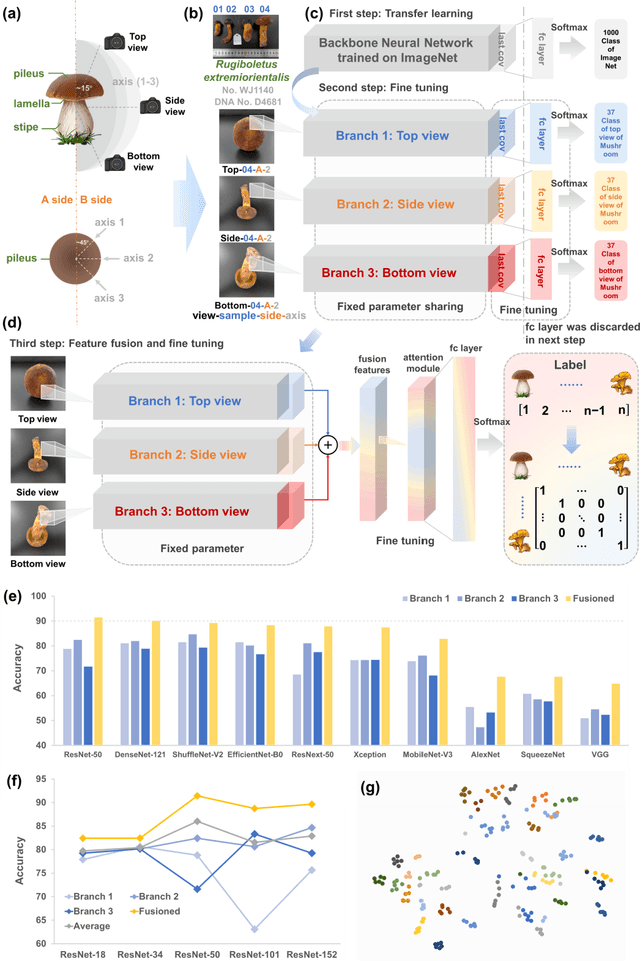
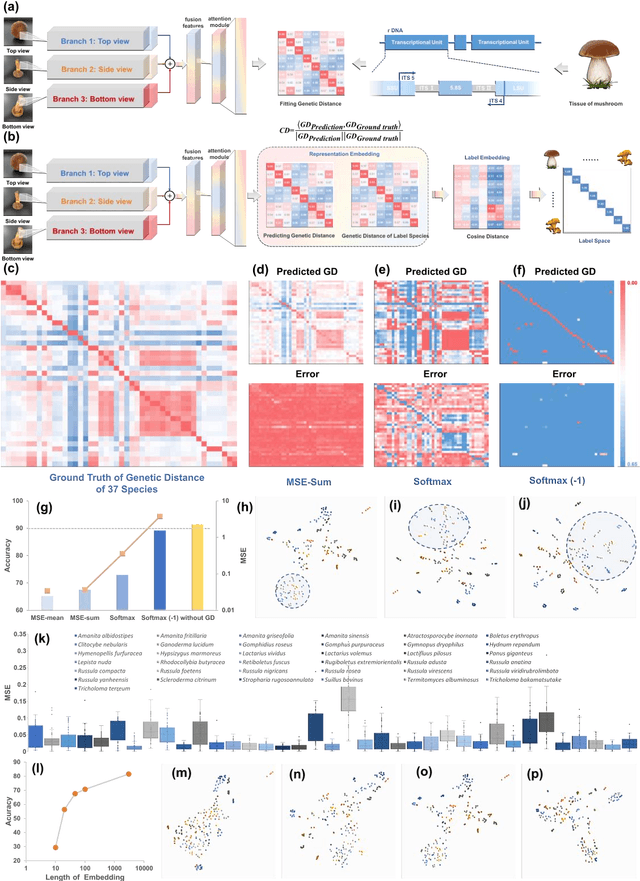
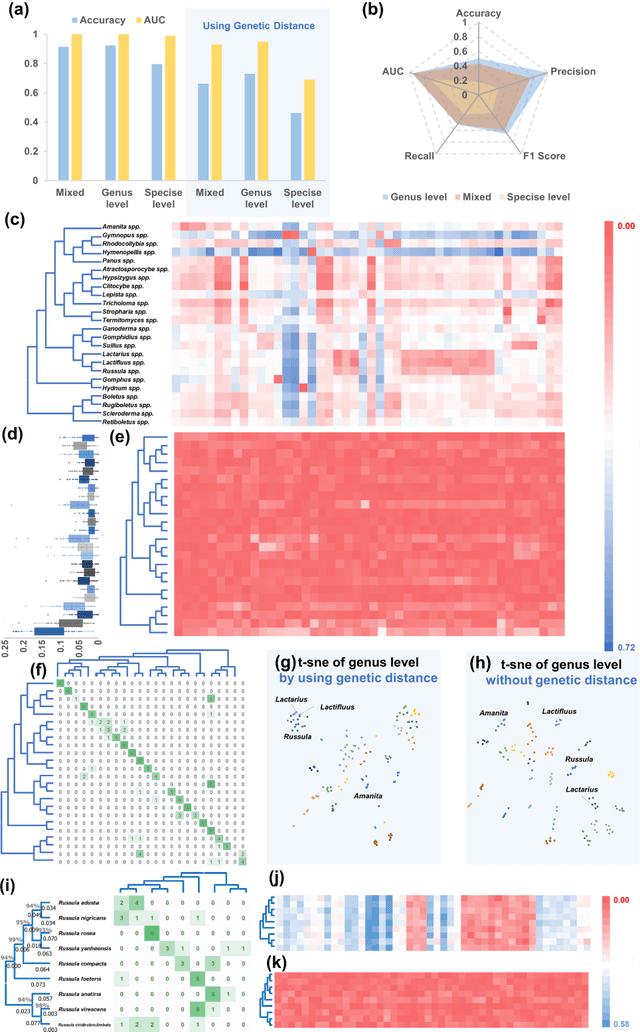
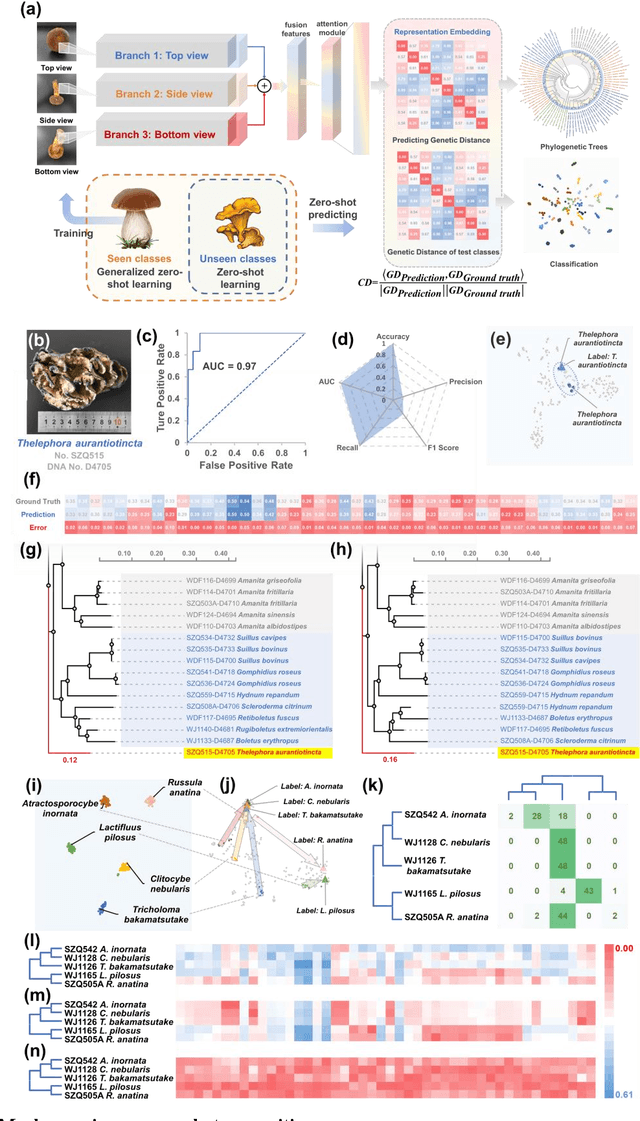
Molecular and morphological characters, as important parts of biological taxonomy, are contradictory but need to be integrated. Organism's image recognition and bioinformatics are emerging and hot problems nowadays but with a gap between them. In this work, a multi-branching recognition framework mediated by genetic information bridges this barrier, which establishes the link between macro-morphology and micro-molecular information of mushrooms. The novel multi-perspective structure is proposed to fuse the feature images from three branching models, which significantly improves the accuracy of recognition by about 10% and up to more than 90%. Further, genetic information is implemented to the mushroom image recognition task by using genetic distance embeddings as the representation space for predicting image distance and species identification. Semantic overfitting of traditional classification tasks and the granularity of fine-grained image recognition are also discussed in depth for the first time. The generalizability of the model was investigated in fine-grained scenarios using zero-shot learning tasks, which could predict the taxonomic and evolutionary information of unseen samples. We presented the first method to map images to DNA, namely used an encoder mapping image to genetic distances, and then decoded DNA through a pre-trained decoder, where the total test accuracy on 37 species for DNA prediction is 87.45%. This study creates a novel recognition framework by systematically studying the mushroom image recognition problem, bridging the gap between macroscopic biological information and microscopic molecular information, which will provide a new reference for intelligent biometrics in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge