Vassili Kovalev
Vertical Federated Learning for Effectiveness, Security, Applicability: A Survey
May 25, 2024



Abstract:Vertical Federated Learning (VFL) is a privacy-preserving distributed learning paradigm where different parties collaboratively learn models using partitioned features of shared samples, without leaking private data. Recent research has shown promising results addressing various challenges in VFL, highlighting its potential for practical applications in cross-domain collaboration. However, the corresponding research is scattered and lacks organization. To advance VFL research, this survey offers a systematic overview of recent developments. First, we provide a history and background introduction, along with a summary of the general training protocol of VFL. We then revisit the taxonomy in recent reviews and analyze limitations in-depth. For a comprehensive and structured discussion, we synthesize recent research from three fundamental perspectives: effectiveness, security, and applicability. Finally, we discuss several critical future research directions in VFL, which will facilitate the developments in this field. We provide a collection of research lists and periodically update them at https://github.com/shentt67/VFL_Survey.
Examining the Capability of GANs to Replace Real Biomedical Images in Classification Models Training
Apr 18, 2019



Abstract:In this paper, we explore the possibility of generating artificial biomedical images that can be used as a substitute for real image datasets in applied machine learning tasks. We are focusing on generation of realistic chest X-ray images as well as on the lymph node histology images using the two recent GAN architectures including DCGAN and PGGAN. The possibility of the use of artificial images instead of real ones for training machine learning models was examined by benchmark classification tasks being solved using conventional and deep learning methods. In particular, a comparison was made by replacing real images with synthetic ones at the model training stage and comparing the prediction results with the ones obtained while training on the real image data. It was found that the drop of classification accuracy caused by such training data substitution ranged between 2.2% and 3.5% for deep learning models and between 5.5% and 13.25% for conventional methods such as LBP + Random Forests.
Influence of Control Parameters and the Size of Biomedical Image Datasets on the Success of Adversarial Attacks
Apr 15, 2019



Abstract:In this paper, we study dependence of the success rate of adversarial attacks to the Deep Neural Networks on the biomedical image type, control parameters, and image dataset size. With this work, we are going to contribute towards accumulation of experimental results on adversarial attacks for the community dealing with biomedical images. The white-box Projected Gradient Descent attacks were examined based on 8 classification tasks and 13 image datasets containing a total of 605,080 chest X-ray and 317,000 histology images of malignant tumors. We concluded that: (1) An increase of the amplitude of perturbation in generating malicious adversarial images leads to a growth of the fraction of successful attacks for the majority of image types examined in this study. (2) Histology images tend to be less sensitive to the growth of amplitude of adversarial perturbations. (3) Percentage of successful attacks is growing with an increase of the number of iterations of the algorithm of generating adversarial perturbations with an asymptotic stabilization. (4) It was found that the success of attacks dropping dramatically when the original confidence of predicting image class exceeds 0.95. (5) The expected dependence of the percentage of successful attacks on the size of image training set was not confirmed.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018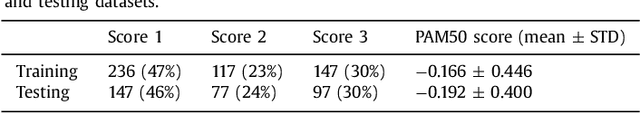
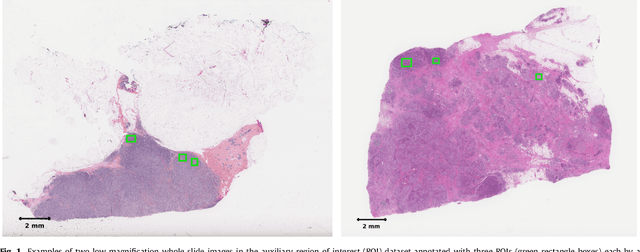
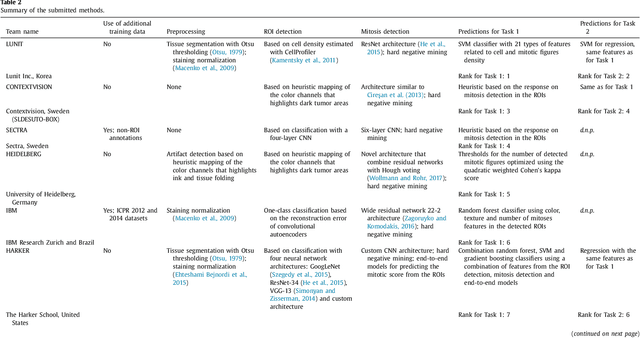
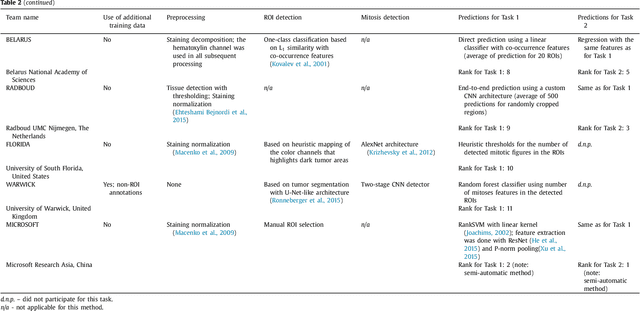
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge