Trent Kyono
To Impute or not to Impute? -- Missing Data in Treatment Effect Estimation
Feb 04, 2022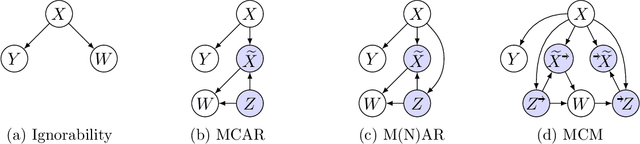
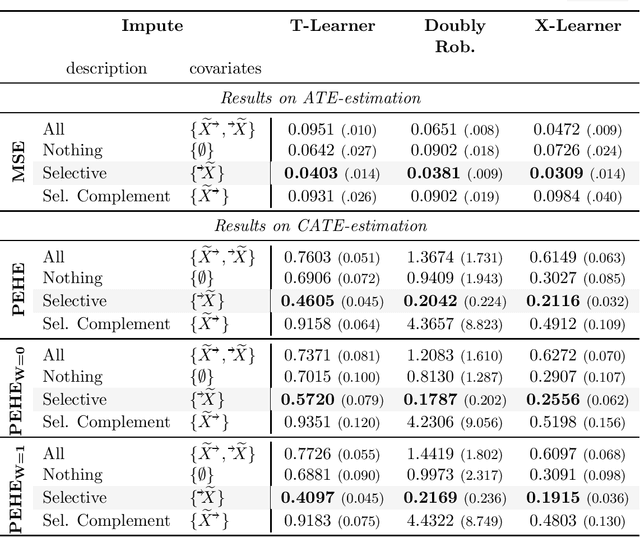
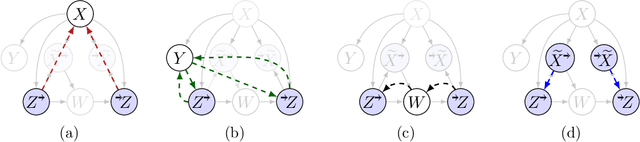
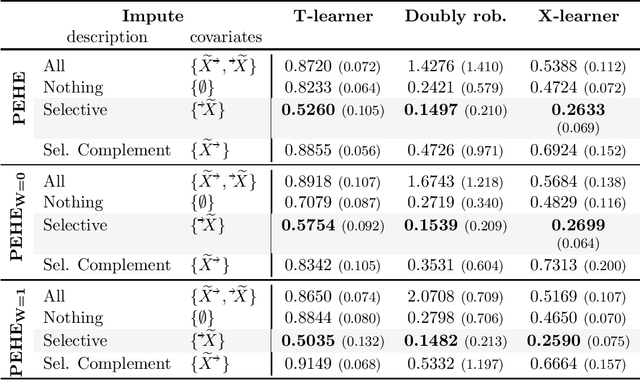
Abstract:Missing data is a systemic problem in practical scenarios that causes noise and bias when estimating treatment effects. This makes treatment effect estimation from data with missingness a particularly tricky endeavour. A key reason for this is that standard assumptions on missingness are rendered insufficient due to the presence of an additional variable, treatment, besides the individual and the outcome. Having a treatment variable introduces additional complexity with respect to why some variables are missing that is not fully explored by previous work. In our work we identify a new missingness mechanism, which we term mixed confounded missingness (MCM), where some missingness determines treatment selection and other missingness is determined by treatment selection. Given MCM, we show that naively imputing all data leads to poor performing treatment effects models, as the act of imputation effectively removes information necessary to provide unbiased estimates. However, no imputation at all also leads to biased estimates, as missingness determined by treatment divides the population in distinct subpopulations, where estimates across these populations will be biased. Our solution is selective imputation, where we use insights from MCM to inform precisely which variables should be imputed and which should not. We empirically demonstrate how various learners benefit from selective imputation compared to other solutions for missing data.
MIRACLE: Causally-Aware Imputation via Learning Missing Data Mechanisms
Nov 04, 2021



Abstract:Missing data is an important problem in machine learning practice. Starting from the premise that imputation methods should preserve the causal structure of the data, we develop a regularization scheme that encourages any baseline imputation method to be causally consistent with the underlying data generating mechanism. Our proposal is a causally-aware imputation algorithm (MIRACLE). MIRACLE iteratively refines the imputation of a baseline by simultaneously modeling the missingness generating mechanism, encouraging imputation to be consistent with the causal structure of the data. We conduct extensive experiments on synthetic and a variety of publicly available datasets to show that MIRACLE is able to consistently improve imputation over a variety of benchmark methods across all three missingness scenarios: at random, completely at random, and not at random.
DECAF: Generating Fair Synthetic Data Using Causally-Aware Generative Networks
Nov 04, 2021



Abstract:Machine learning models have been criticized for reflecting unfair biases in the training data. Instead of solving for this by introducing fair learning algorithms directly, we focus on generating fair synthetic data, such that any downstream learner is fair. Generating fair synthetic data from unfair data - while remaining truthful to the underlying data-generating process (DGP) - is non-trivial. In this paper, we introduce DECAF: a GAN-based fair synthetic data generator for tabular data. With DECAF we embed the DGP explicitly as a structural causal model in the input layers of the generator, allowing each variable to be reconstructed conditioned on its causal parents. This procedure enables inference time debiasing, where biased edges can be strategically removed for satisfying user-defined fairness requirements. The DECAF framework is versatile and compatible with several popular definitions of fairness. In our experiments, we show that DECAF successfully removes undesired bias and - in contrast to existing methods - is capable of generating high-quality synthetic data. Furthermore, we provide theoretical guarantees on the generator's convergence and the fairness of downstream models.
Selecting Treatment Effects Models for Domain Adaptation Using Causal Knowledge
Feb 11, 2021



Abstract:Selecting causal inference models for estimating individualized treatment effects (ITE) from observational data presents a unique challenge since the counterfactual outcomes are never observed. The problem is challenged further in the unsupervised domain adaptation (UDA) setting where we only have access to labeled samples in the source domain, but desire selecting a model that achieves good performance on a target domain for which only unlabeled samples are available. Existing techniques for UDA model selection are designed for the predictive setting. These methods examine discriminative density ratios between the input covariates in the source and target domain and do not factor in the model's predictions in the target domain. Because of this, two models with identical performance on the source domain would receive the same risk score by existing methods, but in reality, have significantly different performance in the test domain. We leverage the invariance of causal structures across domains to propose a novel model selection metric specifically designed for ITE methods under the UDA setting. In particular, we propose selecting models whose predictions of interventions' effects satisfy known causal structures in the target domain. Experimentally, our method selects ITE models that are more robust to covariate shifts on several healthcare datasets, including estimating the effect of ventilation in COVID-19 patients from different geographic locations.
CASTLE: Regularization via Auxiliary Causal Graph Discovery
Sep 28, 2020



Abstract:Regularization improves generalization of supervised models to out-of-sample data. Prior works have shown that prediction in the causal direction (effect from cause) results in lower testing error than the anti-causal direction. However, existing regularization methods are agnostic of causality. We introduce Causal Structure Learning (CASTLE) regularization and propose to regularize a neural network by jointly learning the causal relationships between variables. CASTLE learns the causal directed acyclical graph (DAG) as an adjacency matrix embedded in the neural network's input layers, thereby facilitating the discovery of optimal predictors. Furthermore, CASTLE efficiently reconstructs only the features in the causal DAG that have a causal neighbor, whereas reconstruction-based regularizers suboptimally reconstruct all input features. We provide a theoretical generalization bound for our approach and conduct experiments on a plethora of synthetic and real publicly available datasets demonstrating that CASTLE consistently leads to better out-of-sample predictions as compared to other popular benchmark regularizers.
Improving Model Robustness Using Causal Knowledge
Nov 27, 2019



Abstract:For decades, researchers in fields, such as the natural and social sciences, have been verifying causal relationships and investigating hypotheses that are now well-established or understood as truth. These causal mechanisms are properties of the natural world, and thus are invariant conditions regardless of the collection domain or environment. We show in this paper how prior knowledge in the form of a causal graph can be utilized to guide model selection, i.e., to identify from a set of trained networks the models that are the most robust and invariant to unseen domains. Our method incorporates prior knowledge (which can be incomplete) as a Structural Causal Model (SCM) and calculates a score based on the likelihood of the SCM given the target predictions of a candidate model and the provided input variables. We show on both publicly available and synthetic datasets that our method is able to identify more robust models in terms of generalizability to unseen out-of-distribution test examples and domains where covariates have shifted.
Siamese Survival Analysis with Competing Risks
Aug 16, 2018



Abstract:Survival analysis in the presence of multiple possible adverse events, i.e., competing risks, is a pervasive problem in many industries (healthcare, finance, etc.). Since only one event is typically observed, the incidence of an event of interest is often obscured by other related competing events. This nonidentifiability, or inability to estimate true cause-specific survival curves from empirical data, further complicates competing risk survival analysis. We introduce Siamese Survival Prognosis Network (SSPN), a novel deep learning architecture for estimating personalized risk scores in the presence of competing risks. SSPN circumvents the nonidentifiability problem by avoiding the estimation of cause-specific survival curves and instead determines pairwise concordant time-dependent risks, where longer event times are assigned lower risks. Furthermore, SSPN is able to directly optimize an approximation to the C-discrimination index, rather than relying on well-known metrics which are unable to capture the unique requirements of survival analysis with competing risks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge