Tomas Teijeiro
Cough-E: A multimodal, privacy-preserving cough detection algorithm for the edge
Oct 31, 2024

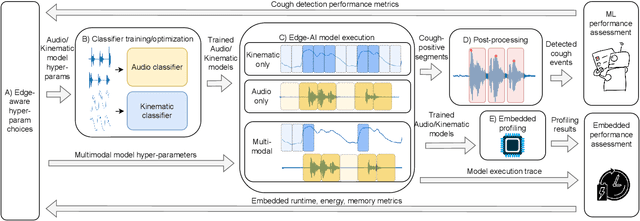

Abstract:Continuous cough monitors can greatly aid doctors in home monitoring and treatment of respiratory diseases. Although many algorithms have been proposed, they still face limitations in data privacy and short-term monitoring. Edge-AI offers a promising solution by processing privacy-sensitive data near the source, but challenges arise in deploying resource-intensive algorithms on constrained devices. From a suitable selection of audio and kinematic signals, our methodology aims at the optimal selection of features via Recursive Feature Elimination with Cross-Validation (RFECV), which exploits the explainability of the selected XGB model. Additionally, it analyzes the use of Mel spectrogram features, instead of the more common MFCC. Moreover, a set of hyperparameters for a multimodal implementation of the classifier is explored. Finally, it evaluates the performance based on clinically relevant event-based metrics. We apply our methodology to develop Cough-E, an energy-efficient, multimodal and edge AI cough detection algorithm. It exploits audio and kinematic data in two distinct classifiers, jointly cooperating for a balanced energy and performance trade-off. We demonstrate that our algorithm can be executed in real-time on an ARM Cortex M33 microcontroller. Cough-E achieves a 70.56\% energy saving when compared to the audio-only approach, at the cost of a 1.26\% relative performance drop, resulting in a 0.78 F1-score. Both Cough-E and the edge-aware model optimization methodology are publicly available as open-source code. This approach demonstrates the benefits of the proposed hardware-aware methodology to enable privacy-preserving cough monitors on the edge, paving the way to efficient cough monitoring.
How to Count Coughs: An Event-Based Framework for Evaluating Automatic Cough Detection Algorithm Performance
Jun 03, 2024Abstract:Chronic cough disorders are widespread and challenging to assess because they rely on subjective patient questionnaires about cough frequency. Wearable devices running Machine Learning (ML) algorithms are promising for quantifying daily coughs, providing clinicians with objective metrics to track symptoms and evaluate treatments. However, there is a mismatch between state-of-the-art metrics for cough counting algorithms and the information relevant to clinicians. Most works focus on distinguishing cough from non-cough samples, which does not directly provide clinically relevant outcomes such as the number of cough events or their temporal patterns. In addition, typical metrics such as specificity and accuracy can be biased by class imbalance. We propose using event-based evaluation metrics aligned with clinical guidelines on significant cough counting endpoints. We use an ML classifier to illustrate the shortcomings of traditional sample-based accuracy measurements, highlighting their variance due to dataset class imbalance and sample window length. We also present an open-source event-based evaluation framework to test algorithm performance in identifying cough events and rejecting false positives. We provide examples and best practice guidelines in event-based cough counting as a necessary first step to assess algorithm performance with clinical relevance.
Acoustical Features as Knee Health Biomarkers: A Critical Analysis
May 23, 2024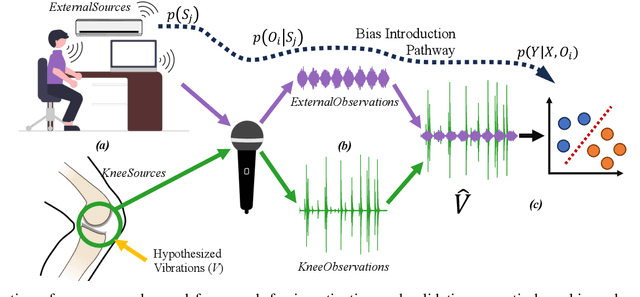
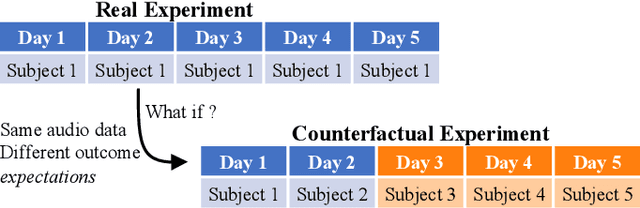
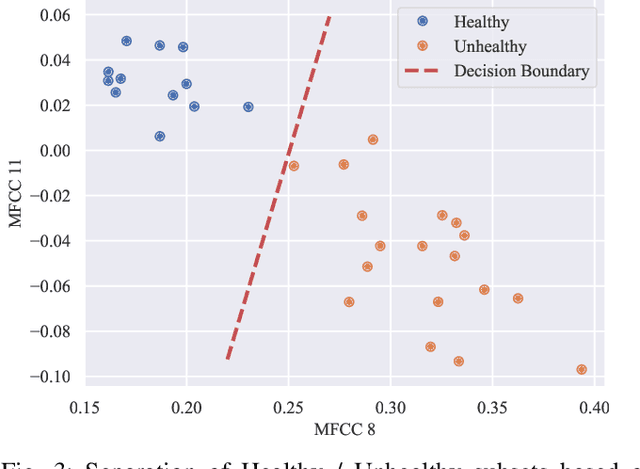
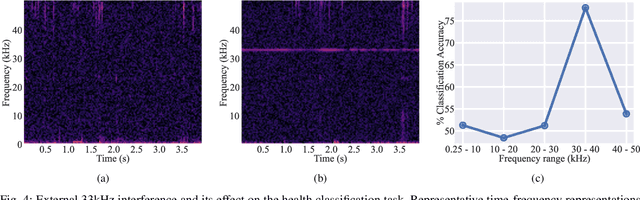
Abstract:Acoustical knee health assessment has long promised an alternative to clinically available medical imaging tools, but this modality has yet to be adopted in medical practice. The field is currently led by machine learning models processing acoustical features, which have presented promising diagnostic performances. However, these methods overlook the intricate multi-source nature of audio signals and the underlying mechanisms at play. By addressing this critical gap, the present paper introduces a novel causal framework for validating knee acoustical features. We argue that current machine learning methodologies for acoustical knee diagnosis lack the required assurances and thus cannot be used to classify acoustic features as biomarkers. Our framework establishes a set of essential theoretical guarantees necessary to validate this claim. We apply our methodology to three real-world experiments investigating the effect of researchers' expectations, the experimental protocol and the wearable employed sensor. This investigation reveals latent issues such as underlying shortcut learning and performance inflation. This study is the first independent result reproduction study in the field of acoustical knee health evaluation. We conclude with actionable insights from our findings, offering valuable guidance to navigate these crucial limitations in future research.
Machine Learning Discovery of Optimal Quadrature Rules for Isogeometric Analysis
Apr 04, 2023Abstract:We propose the use of machine learning techniques to find optimal quadrature rules for the construction of stiffness and mass matrices in isogeometric analysis (IGA). We initially consider 1D spline spaces of arbitrary degree spanned over uniform and non-uniform knot sequences, and then the generated optimal rules are used for integration over higher-dimensional spaces using tensor product sense. The quadrature rule search is posed as an optimization problem and solved by a machine learning strategy based on gradient-descent. However, since the optimization space is highly non-convex, the success of the search strongly depends on the number of quadrature points and the parameter initialization. Thus, we use a dynamic programming strategy that initializes the parameters from the optimal solution over the spline space with a lower number of knots. With this method, we found optimal quadrature rules for spline spaces when using IGA discretizations with up to 50 uniform elements and polynomial degrees up to 8, showing the generality of the approach in this scenario. For non-uniform partitions, the method also finds an optimal rule in a reasonable number of test cases. We also assess the generated optimal rules in two practical case studies, namely, the eigenvalue problem of the Laplace operator and the eigenfrequency analysis of freeform curved beams, where the latter problem shows the applicability of the method to curved geometries. In particular, the proposed method results in savings with respect to traditional Gaussian integration of up to 44% in 1D, 68% in 2D, and 82% in 3D spaces.
Combining General and Personalized Models for Epilepsy Detection with Hyperdimensional Computing
Mar 26, 2023



Abstract:Epilepsy is a chronic neurological disorder with a significant prevalence. However, there is still no adequate technological support to enable epilepsy detection and continuous outpatient monitoring in everyday life. Hyperdimensional (HD) computing is an interesting alternative for wearable devices, characterized by a much simpler learning process and also lower memory requirements. In this work, we demonstrate a few additional aspects in which HD computing, and the way its models are built and stored, can be used for further understanding, comparing, and creating more advanced machine learning models for epilepsy detection. These possibilities are not feasible with other state-of-the-art models, such as random forests or neural networks. We compare inter-subject similarity of models per different classes (seizure and non-seizure), then study the process of creation of generalized models from personalized ones, and in the end, how to combine personalized and generalized models to create hybrid models. This results in improved epilepsy detection performance. We also tested knowledge transfer between models created on two different datasets. Finally, all those examples could be highly interesting not only from an engineering perspective to create better models for wearables, but also from a neurological perspective to better understand individual epilepsy patterns.
Importance of methodological choices in data manipulation for validating epileptic seizure detection models
Feb 21, 2023Abstract:Epilepsy is a chronic neurological disorder that affects a significant portion of the human population and imposes serious risks in the daily life of patients. Despite advances in machine learning and IoT, small, nonstigmatizing wearable devices for continuous monitoring and detection in outpatient environments are not yet available. Part of the reason is the complexity of epilepsy itself, including highly imbalanced data, multimodal nature, and very subject-specific signatures. However, another problem is the heterogeneity of methodological approaches in research, leading to slower progress, difficulty comparing results, and low reproducibility. Therefore, this article identifies a wide range of methodological decisions that must be made and reported when training and evaluating the performance of epilepsy detection systems. We characterize the influence of individual choices using a typical ensemble random-forest model and the publicly available CHB-MIT database, providing a broader picture of each decision and giving good-practice recommendations, based on our experience, where possible.
A Semi-Supervised Algorithm for Improving the Consistency of Crowdsourced Datasets: The COVID-19 Case Study on Respiratory Disorder Classification
Sep 09, 2022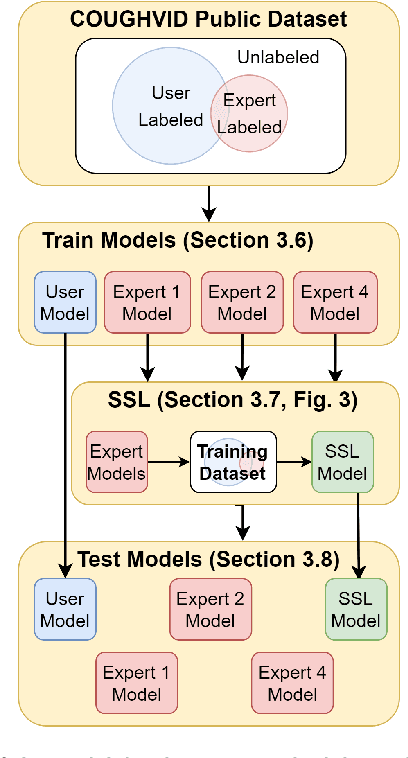

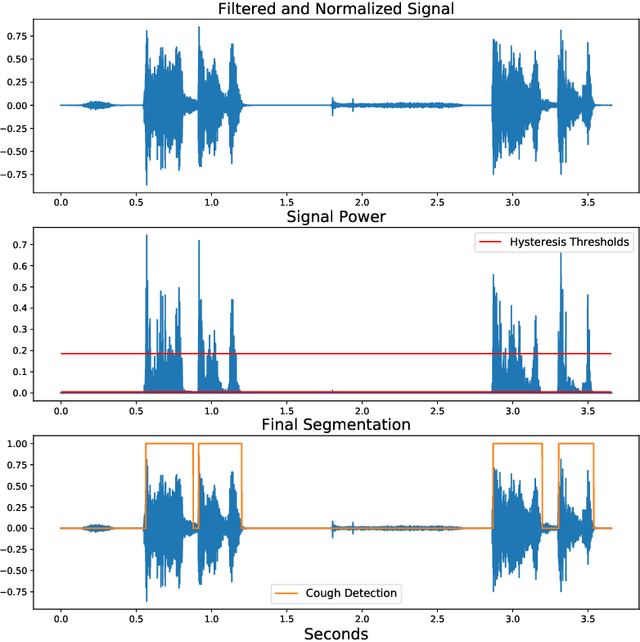
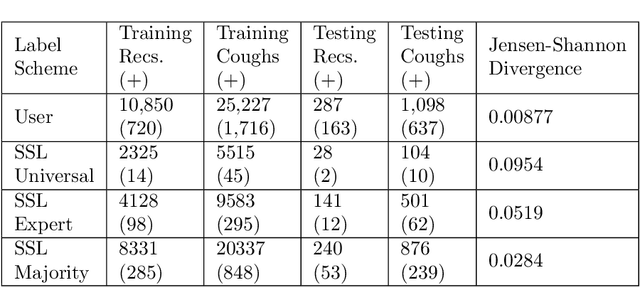
Abstract:Cough audio signal classification is a potentially useful tool in screening for respiratory disorders, such as COVID-19. Since it is dangerous to collect data from patients with such contagious diseases, many research teams have turned to crowdsourcing to quickly gather cough sound data, as it was done to generate the COUGHVID dataset. The COUGHVID dataset enlisted expert physicians to diagnose the underlying diseases present in a limited number of uploaded recordings. However, this approach suffers from potential mislabeling of the coughs, as well as notable disagreement between experts. In this work, we use a semi-supervised learning (SSL) approach to improve the labeling consistency of the COUGHVID dataset and the robustness of COVID-19 versus healthy cough sound classification. First, we leverage existing SSL expert knowledge aggregation techniques to overcome the labeling inconsistencies and sparsity in the dataset. Next, our SSL approach is used to identify a subsample of re-labeled COUGHVID audio samples that can be used to train or augment future cough classification models. The consistency of the re-labeled data is demonstrated in that it exhibits a high degree of class separability, 3x higher than that of the user-labeled data, despite the expert label inconsistency present in the original dataset. Furthermore, the spectral differences in the user-labeled audio segments are amplified in the re-labeled data, resulting in significantly different power spectral densities between healthy and COVID-19 coughs, which demonstrates both the increased consistency of the new dataset and its explainability from an acoustic perspective. Finally, we demonstrate how the re-labeled dataset can be used to train a cough classifier. This SSL approach can be used to combine the medical knowledge of several experts to improve the database consistency for any diagnostic classification task.
Many-to-One Knowledge Distillation of Real-Time Epileptic Seizure Detection for Low-Power Wearable Internet of Things Systems
Jul 20, 2022



Abstract:Integrating low-power wearable Internet of Things (IoT) systems into routine health monitoring is an ongoing challenge. Recent advances in the computation capabilities of wearables make it possible to target complex scenarios by exploiting multiple biosignals and using high-performance algorithms, such as Deep Neural Networks (DNNs). There is, however, a trade-off between performance of the algorithms and the low-power requirements of IoT platforms with limited resources. Besides, physically larger and multi-biosignal-based wearables bring significant discomfort to the patients. Consequently, reducing power consumption and discomfort is necessary for patients to use IoT devices continuously during everyday life. To overcome these challenges, in the context of epileptic seizure detection, we propose a many-to-one signals knowledge distillation approach targeting single-biosignal processing in IoT wearable systems. The starting point is to get a highly-accurate multi-biosignal DNN, then apply our approach to develop a single-biosignal DNN solution for IoT systems that achieves an accuracy comparable to the original multi-biosignal DNN. To assess the practicality of our approach to real-life scenarios, we perform a comprehensive simulation experiment analysis on several state-of-the-art edge computing platforms, such as Kendryte K210 and Raspberry Pi Zero.
Event-based sampled ECG morphology reconstruction through self-similarity
Jul 05, 2022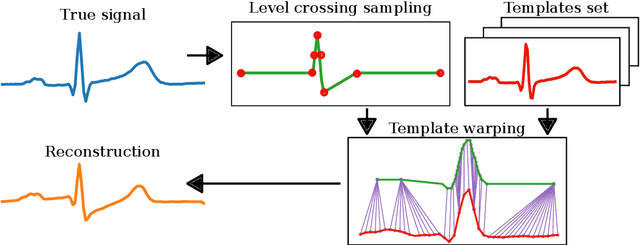


Abstract:Background and Objective: Event-based analog-to-digital converters allow for sparse bio-signal acquisition, enabling local sub-Nyquist sampling frequency. However, aggressive event selection can cause the loss of important bio-markers, not recoverable with standard interpolation techniques. In this work, we leverage the self-similarity of the electrocardiogram (ECG) signal to recover missing features in event-based sampled ECG signals, dynamically selecting patient-representative templates together with a novel dynamic time warping algorithm to infer the morphology of event-based sampled heartbeats. Methods: We acquire a set of uniformly sampled heartbeats and use a graph-based clustering algorithm to define representative templates for the patient. Then, for each event-based sampled heartbeat, we select the morphologically nearest template, and we then reconstruct the heartbeat with piece-wise linear deformations of the selected template, according to a novel dynamic time warping algorithm that matches events to template segments. Results: Synthetic tests on a standard normal sinus rhythm dataset, composed of approximately 1.8 million normal heartbeats, show a big leap in performance with respect to standard resampling techniques. In particular (when compared to classic linear resampling), we show an improvement in P-wave detection of up to 10 times, an improvement in T-wave detection of up to three times, and a 30\% improvement in the dynamic time warping morphological distance. Conclusion: In this work, we have developed an event-based processing pipeline that leverages signal self-similarity to reconstruct event-based sampled ECG signals. Synthetic tests show clear advantages over classical resampling techniques.
HDTorch: Accelerating Hyperdimensional Computing with GP-GPUs for Design Space Exploration
Jun 09, 2022



Abstract:HyperDimensional Computing (HDC) as a machine learning paradigm is highly interesting for applications involving continuous, semi-supervised learning for long-term monitoring. However, its accuracy is not yet on par with other Machine Learning (ML) approaches. Frameworks enabling fast design space exploration to find practical algorithms are necessary to make HD computing competitive with other ML techniques. To this end, we introduce HDTorch, an open-source, PyTorch-based HDC library with CUDA extensions for hypervector operations. We demonstrate HDTorch's utility by analyzing four HDC benchmark datasets in terms of accuracy, runtime, and memory consumption, utilizing both classical and online HD training methodologies. We demonstrate average (training)/inference speedups of (111x/68x)/87x for classical/online HD, respectively. Moreover, we analyze the effects of varying hyperparameters on runtime and accuracy. Finally, we demonstrate how HDTorch enables exploration of HDC strategies applied to large, real-world datasets. We perform the first-ever HD training and inference analysis of the entirety of the CHB-MIT EEG epilepsy database. Results show that the typical approach of training on a subset of the data does not necessarily generalize to the entire dataset, an important factor when developing future HD models for medical wearable devices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge