Tianshuo Zhou
Sampling-based Continuous Optimization with Coupled Variables for RNA Design
Dec 11, 2024Abstract:The task of RNA design given a target structure aims to find a sequence that can fold into that structure. It is a computationally hard problem where some version(s) have been proven to be NP-hard. As a result, heuristic methods such as local search have been popular for this task, but by only exploring a fixed number of candidates. They can not keep up with the exponential growth of the design space, and often perform poorly on longer and harder-to-design structures. We instead formulate these discrete problems as continuous optimization, which starts with a distribution over all possible candidate sequences, and uses gradient descent to improve the expectation of an objective function. We define novel distributions based on coupled variables to rule out invalid sequences given the target structure and to model the correlation between nucleotides. To make it universally applicable to any objective function, we use sampling to approximate the expected objective function, to estimate the gradient, and to select the final candidate. Compared to the state-of-the-art methods, our work consistently outperforms them in key metrics such as Boltzmann probability, ensemble defect, and energy gap, especially on long and hard-to-design puzzles in the Eterna100 benchmark. Our code is available at: http://github.com/weiyutang1010/ncrna_design.
GUI-WORLD: A Dataset for GUI-oriented Multimodal LLM-based Agents
Jun 16, 2024

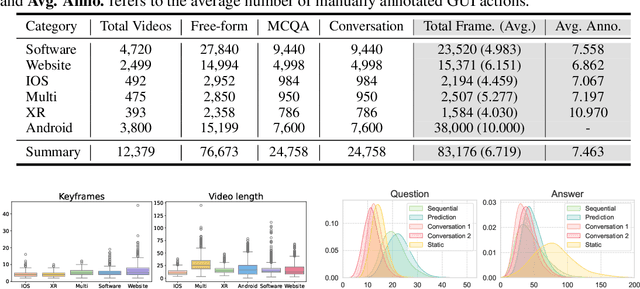

Abstract:Recently, Multimodal Large Language Models (MLLMs) have been used as agents to control keyboard and mouse inputs by directly perceiving the Graphical User Interface (GUI) and generating corresponding code. However, current agents primarily exhibit excellent understanding capabilities in static environments and are predominantly applied in relatively simple domains, such as Web or mobile interfaces. We argue that a robust GUI agent should be capable of perceiving temporal information on the GUI, including dynamic Web content and multi-step tasks. Additionally, it should possess a comprehensive understanding of various GUI scenarios, including desktop software and multi-window interactions. To this end, this paper introduces a new dataset, termed GUI-World, which features meticulously crafted Human-MLLM annotations, extensively covering six GUI scenarios and eight types of GUI-oriented questions in three formats. We evaluate the capabilities of current state-of-the-art MLLMs, including ImageLLMs and VideoLLMs, in understanding various types of GUI content, especially dynamic and sequential content. Our findings reveal that ImageLLMs struggle with dynamic GUI content without manually annotated keyframes or operation history. On the other hand, VideoLLMs fall short in all GUI-oriented tasks given the sparse GUI video dataset. Based on GUI-World, we take the initial step of leveraging a fine-tuned VideoLLM as a GUI agent, demonstrating an improved understanding of various GUI tasks. However, due to the limitations in the performance of base LLMs, we conclude that using VideoLLMs as GUI agents remains a significant challenge. We believe our work provides valuable insights for future research in dynamic GUI content understanding. The code and dataset are publicly available at our project homepage: https://gui-world.github.io/.
Messenger and Non-Coding RNA Design via Expected Partition Function and Continuous Optimization
Dec 29, 2023



Abstract:The tasks of designing messenger RNAs and non-coding RNAs are discrete optimization problems, and several versions of these problems are NP-hard. As an alternative to commonly used local search methods, we formulate these problems as continuous optimization and develop a general framework for this optimization based on a new concept of "expected partition function". The basic idea is to start with a distribution over all possible candidate sequences, and extend the objective function from a sequence to a distribution. We then use gradient descent-based optimization methods to improve the extended objective function, and the distribution will gradually shrink towards a one-hot sequence (i.e., a single sequence). We consider two important case studies within this framework, the mRNA design problem optimizing for partition function (i.e., ensemble free energy) and the non-coding RNA design problem optimizing for conditional (i.e., Boltzmann) probability. In both cases, our approach demonstrate promising preliminary results. We make our code available at https://github.com/KuNyaa/RNA_Design_codebase.
Unlock Multi-Modal Capability of Dense Retrieval via Visual Module Plugin
Oct 21, 2023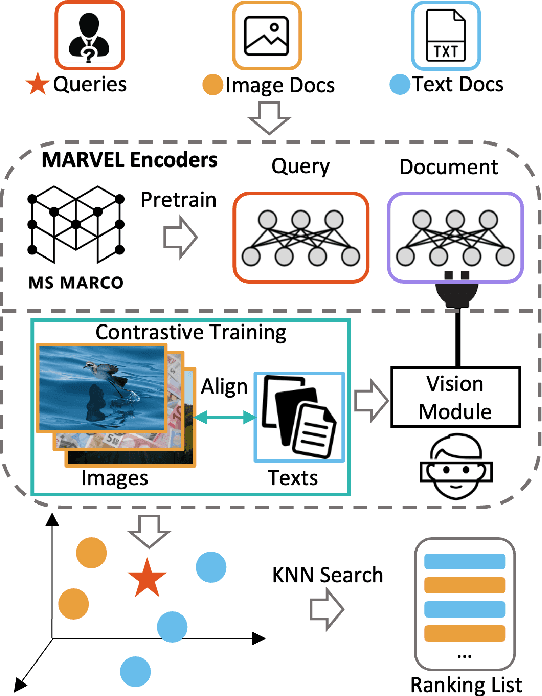
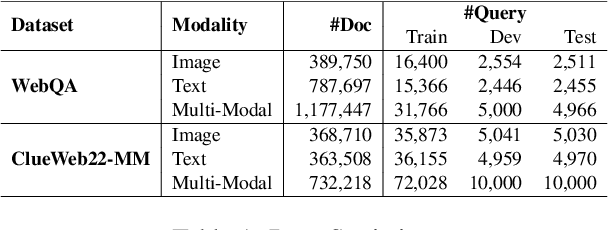
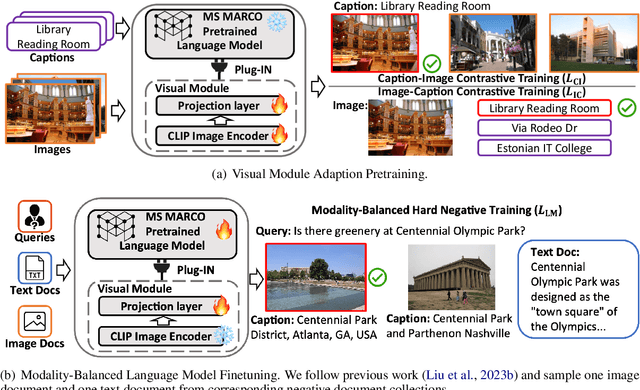
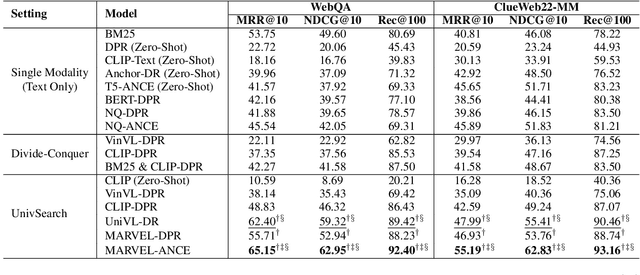
Abstract:This paper proposes Multi-modAl Retrieval model via Visual modulE pLugin (MARVEL) to learn an embedding space for queries and multi-modal documents to conduct retrieval. MARVEL encodes queries and multi-modal documents with a unified encoder model, which helps to alleviate the modality gap between images and texts. Specifically, we enable the image understanding ability of a well-trained dense retriever, T5-ANCE, by incorporating the image features encoded by the visual module as its inputs. To facilitate the multi-modal retrieval tasks, we build the ClueWeb22-MM dataset based on the ClueWeb22 dataset, which regards anchor texts as queries, and exact the related texts and image documents from anchor linked web pages. Our experiments show that MARVEL significantly outperforms the state-of-the-art methods on the multi-modal retrieval dataset WebQA and ClueWeb22-MM. Our further analyses show that the visual module plugin method is tailored to enable the image understanding ability for an existing dense retrieval model. Besides, we also show that the language model has the ability to extract image semantics from image encoders and adapt the image features in the input space of language models. All codes are available at https://github.com/OpenMatch/MARVEL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge