Tiago Azevedo
A Monosemantic Attribution Framework for Stable Interpretability in Clinical Neuroscience Large Language Models
Jan 25, 2026Abstract:Interpretability remains a key challenge for deploying large language models (LLMs) in clinical settings such as Alzheimer's disease progression diagnosis, where early and trustworthy predictions are essential. Existing attribution methods exhibit high inter-method variability and unstable explanations due to the polysemantic nature of LLM representations, while mechanistic interpretability approaches lack direct alignment with model inputs and outputs and do not provide explicit importance scores. We introduce a unified interpretability framework that integrates attributional and mechanistic perspectives through monosemantic feature extraction. By constructing a monosemantic embedding space at the level of an LLM layer and optimizing the framework to explicitly reduce inter-method variability, our approach produces stable input-level importance scores and highlights salient features via a decompressed representation of the layer of interest, advancing the safe and trustworthy application of LLMs in cognitive health and neurodegenerative disease.
Unified Generative Latent Representation for Functional Brain Graphs
Nov 06, 2025Abstract:Functional brain graphs are often characterized with separate graph-theoretic or spectral descriptors, overlooking how these properties covary and partially overlap across brains and conditions. We anticipate that dense, weighted functional connectivity graphs occupy a low-dimensional latent geometry along which both topological and spectral structures display graded variations. Here, we estimated this unified graph representation and enabled generation of dense functional brain graphs through a graph transformer autoencoder with latent diffusion, with spectral geometry providing an inductive bias to guide learning. This geometry-aware latent representation, although unsupervised, meaningfully separated working-memory states and decoded visual stimuli, with performance further enhanced by incorporating neural dynamics. From the diffusion modeled distribution, we were able to sample biologically plausible and structurally grounded synthetic dense graphs.
Artificial Intelligence-powered fossil shark tooth identification: Unleashing the potential of Convolutional Neural Networks
May 07, 2024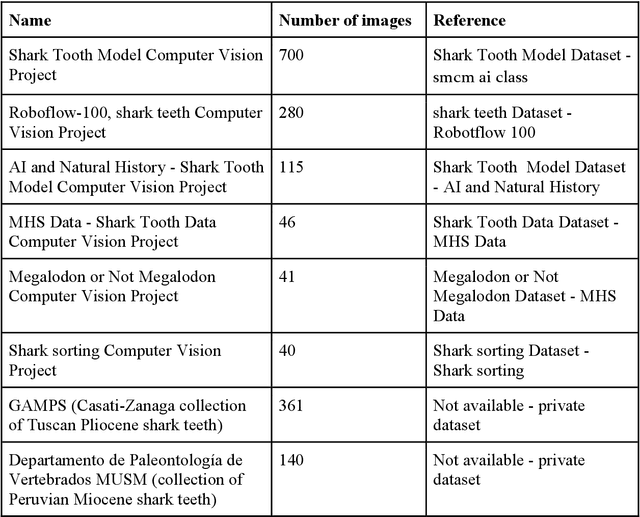

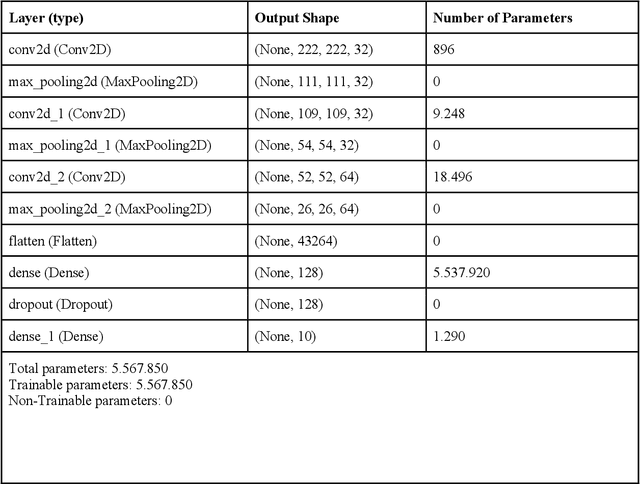
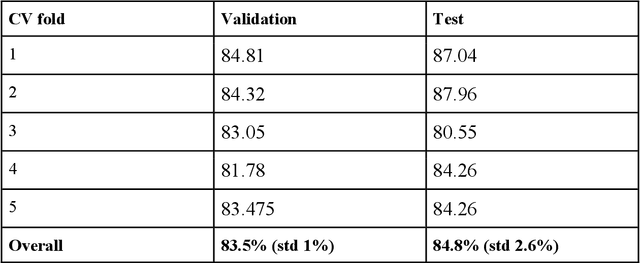
Abstract:All fields of knowledge are being impacted by Artificial Intelligence. In particular, the Deep Learning paradigm enables the development of data analysis tools that support subject matter experts in a variety of sectors, from physics up to the recognition of ancient languages. Palaeontology is now observing this trend as well. This study explores the capability of Convolutional Neural Networks (CNNs), a particular class of Deep Learning algorithms specifically crafted for computer vision tasks, to classify images of isolated fossil shark teeth gathered from online datasets as well as from the authors$'$ experience on Peruvian Miocene and Italian Pliocene fossil assemblages. The shark taxa that are included in the final, composite dataset (which consists of more than one thousand images) are representative of both extinct and extant genera, namely, Carcharhinus, Carcharias, Carcharocles, Chlamydoselachus, Cosmopolitodus, Galeocerdo, Hemipristis, Notorynchus, Prionace and Squatina. We developed a CNN, named SharkNet-X, specifically tailored on our recognition task, reaching a 5-fold cross validated mean accuracy of 0.85 to identify images containing a single shark tooth. Furthermore, we elaborated a visualization of the features extracted from images using the last dense layer of the CNN, achieved through the application of the clustering technique t-SNE. In addition, in order to understand and explain the behaviour of the CNN while giving a paleontological point of view on the results, we introduced the explainability method SHAP. To the best of our knowledge, this is the first instance in which this method is applied to the field of palaeontology. The main goal of this work is to showcase how Deep Learning techniques can aid in identifying isolated fossil shark teeth, paving the way for developing new information tools for automating the recognition and classification of fossils.
An Underexplored Dilemma between Confidence and Calibration in Quantized Neural Networks
Dec 02, 2021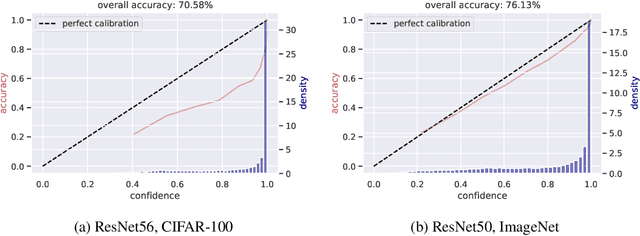
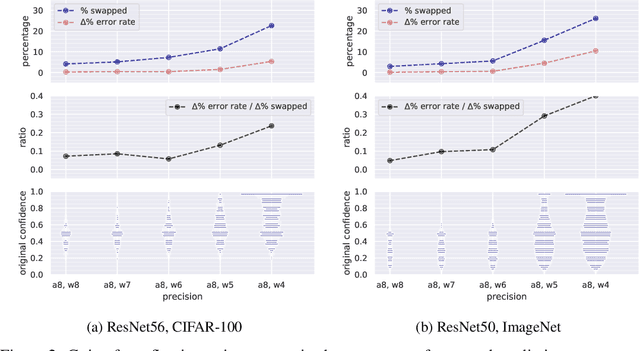
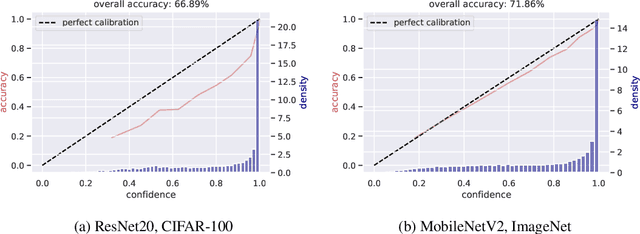
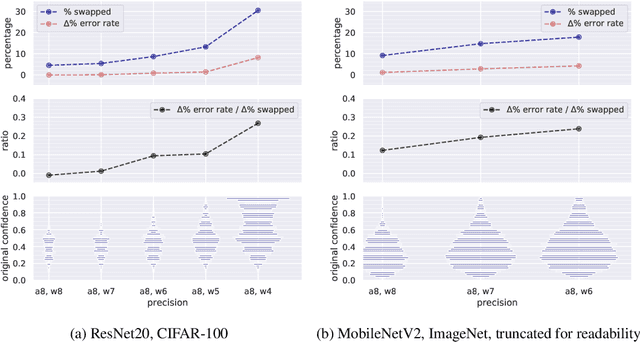
Abstract:Modern convolutional neural networks (CNNs) are known to be overconfident in terms of their calibration on unseen input data. That is to say, they are more confident than they are accurate. This is undesirable if the probabilities predicted are to be used for downstream decision making. When considering accuracy, CNNs are also surprisingly robust to compression techniques, such as quantization, which aim to reduce computational and memory costs. We show that this robustness can be partially explained by the calibration behavior of modern CNNs, and may be improved with overconfidence. This is due to an intuitive result: low confidence predictions are more likely to change post-quantization, whilst being less accurate. High confidence predictions will be more accurate, but more difficult to change. Thus, a minimal drop in post-quantization accuracy is incurred. This presents a potential conflict in neural network design: worse calibration from overconfidence may lead to better robustness to quantization. We perform experiments applying post-training quantization to a variety of CNNs, on the CIFAR-100 and ImageNet datasets.
On Efficient Uncertainty Estimation for Resource-Constrained Mobile Applications
Dec 01, 2021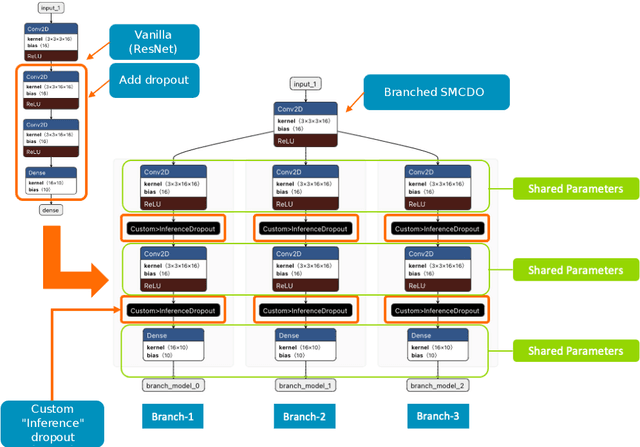
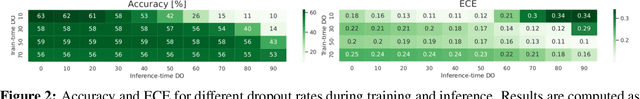
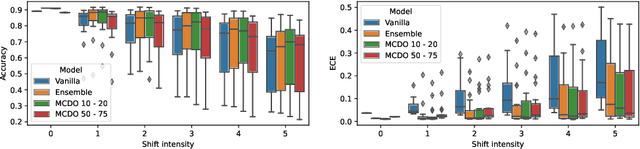
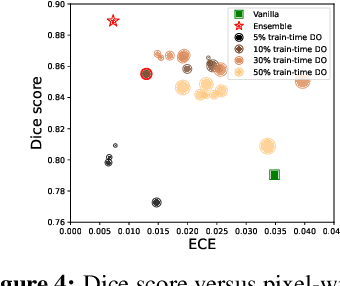
Abstract:Deep neural networks have shown great success in prediction quality while reliable and robust uncertainty estimation remains a challenge. Predictive uncertainty supplements model predictions and enables improved functionality of downstream tasks including embedded and mobile applications, such as virtual reality, augmented reality, sensor fusion, and perception. These applications often require a compromise in complexity to obtain uncertainty estimates due to very limited memory and compute resources. We tackle this problem by building upon Monte Carlo Dropout (MCDO) models using the Axolotl framework; specifically, we diversify sampled subnetworks, leverage dropout patterns, and use a branching technique to improve predictive performance while maintaining fast computations. We conduct experiments on (1) a multi-class classification task using the CIFAR10 dataset, and (2) a more complex human body segmentation task. Our results show the effectiveness of our approach by reaching close to Deep Ensemble prediction quality and uncertainty estimation, while still achieving faster inference on resource-limited mobile platforms.
Towards Efficient Point Cloud Graph Neural Networks Through Architectural Simplification
Aug 13, 2021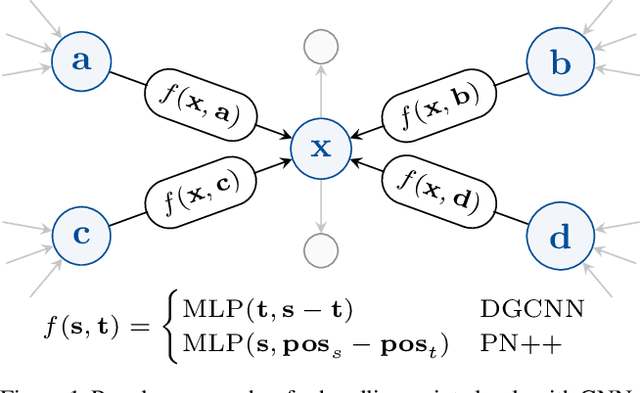
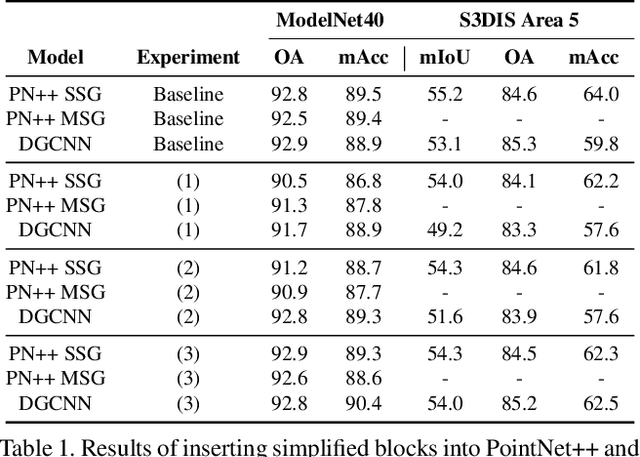
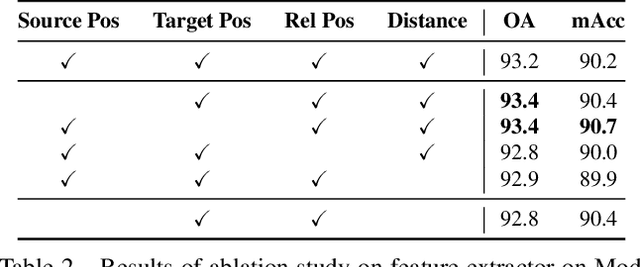
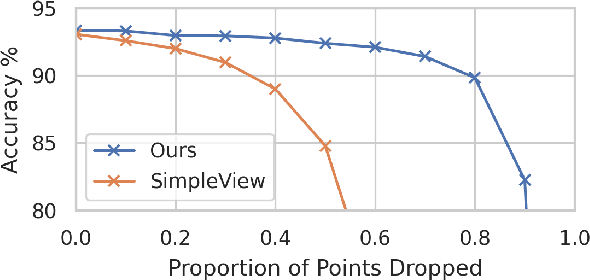
Abstract:In recent years graph neural network (GNN)-based approaches have become a popular strategy for processing point cloud data, regularly achieving state-of-the-art performance on a variety of tasks. To date, the research community has primarily focused on improving model expressiveness, with secondary thought given to how to design models that can run efficiently on resource constrained mobile devices including smartphones or mixed reality headsets. In this work we make a step towards improving the efficiency of these models by making the observation that these GNN models are heavily limited by the representational power of their first, feature extracting, layer. We find that it is possible to radically simplify these models so long as the feature extraction layer is retained with minimal degradation to model performance; further, we discover that it is possible to improve performance overall on ModelNet40 and S3DIS by improving the design of the feature extractor. Our approach reduces memory consumption by 20$\times$ and latency by up to 9.9$\times$ for graph layers in models such as DGCNN; overall, we achieve speed-ups of up to 4.5$\times$ and peak memory reductions of 72.5%.
Stochastic-YOLO: Efficient Probabilistic Object Detection under Dataset Shifts
Sep 07, 2020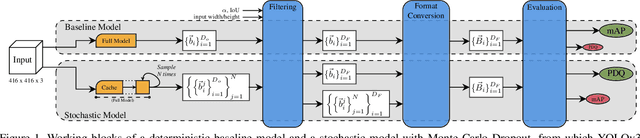
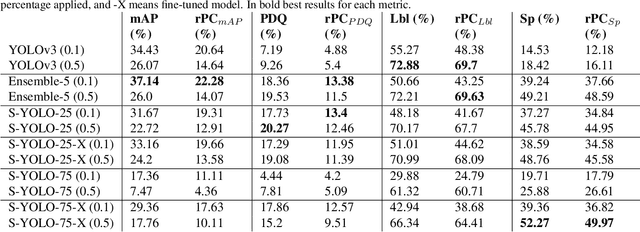
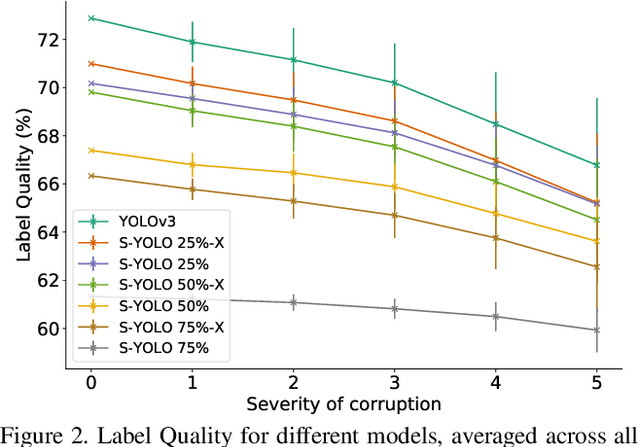
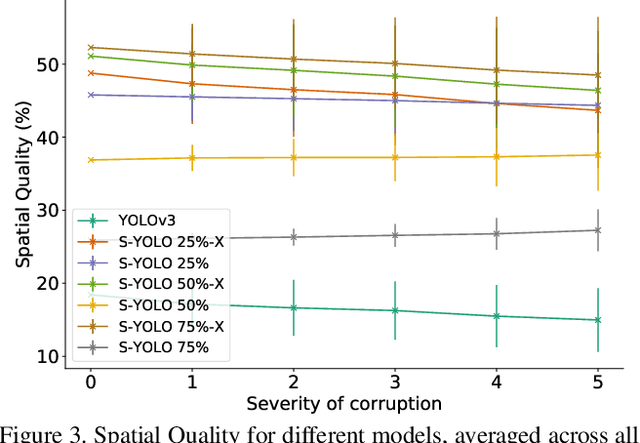
Abstract:In image classification tasks, the evaluation of models' robustness to increased dataset shifts with a probabilistic framework is very well studied. However, Object Detection (OD) tasks pose other challenges for uncertainty estimation and evaluation. For example, one needs to evaluate both the quality of the label uncertainty (i.e., what?) and spatial uncertainty (i.e., where?) for a given bounding box, but that evaluation cannot be performed with more traditional average precision metrics (e.g., mAP). In this paper, we adapt the well-established YOLOv3 architecture to generate uncertainty estimations by introducing stochasticity in the form of Monte Carlo Dropout (MC-Drop), and evaluate it across different levels of dataset shift. We call this novel architecture Stochastic-YOLO, and provide an efficient implementation to effectively reduce the burden of the MC-Drop sampling mechanism at inference time. Finally, we provide some sensitivity analyses, while arguing that Stochastic-YOLO is a sound approach that improves different components of uncertainty estimations, in particular spatial uncertainties.
Towards a predictive spatio-temporal representation of brain data
Feb 29, 2020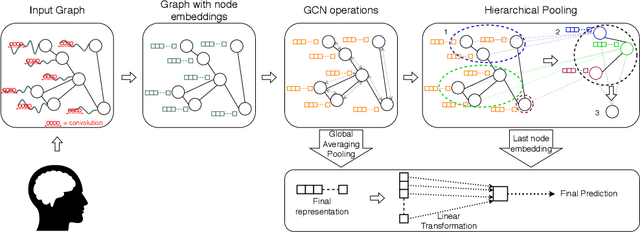
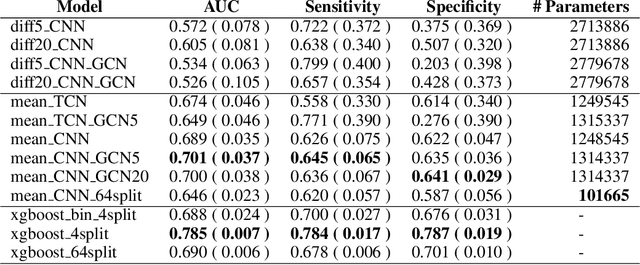
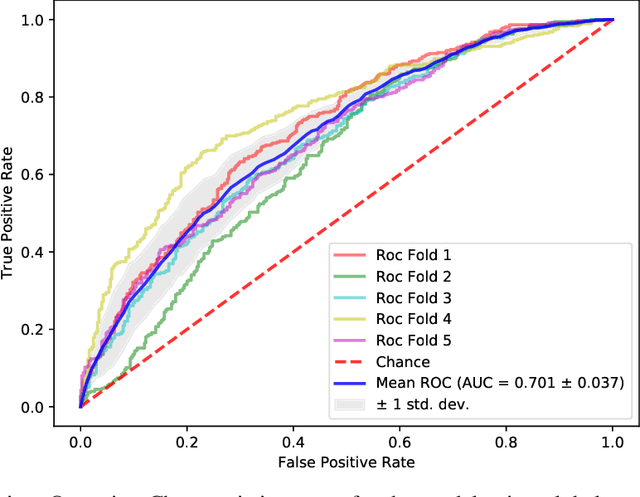
Abstract:The characterisation of the brain as a "connectome", in which the connections are represented by correlational values across timeseries and as summary measures derived from graph theory analyses, has been very popular in the last years. However, although this representation has advanced our understanding of the brain function, it may represent an oversimplified model. This is because the typical fMRI datasets are constituted by complex and highly heterogeneous timeseries that vary across space (i.e., location of brain regions). We compare various modelling techniques from deep learning and geometric deep learning to pave the way for future research in effectively leveraging the rich spatial and temporal domains of typical fMRI datasets, as well as of other similar datasets. As a proof-of-concept, we compare our approaches in the homogeneous and publicly available Human Connectome Project (HCP) dataset on a supervised binary classification task. We hope that our methodological advances relative to previous "connectomic" measures can ultimately be clinically and computationally relevant by leading to a more nuanced understanding of the brain dynamics in health and disease. Such understanding of the brain can fundamentally reduce the constant specialised clinical expertise in order to accurately understand brain variability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge