Thomas P. Quinn
Data Augmentation for Compositional Data: Advancing Predictive Models of the Microbiome
May 20, 2022
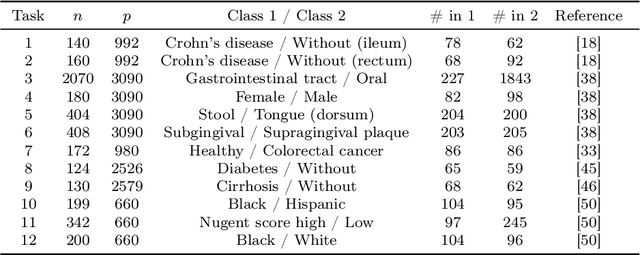
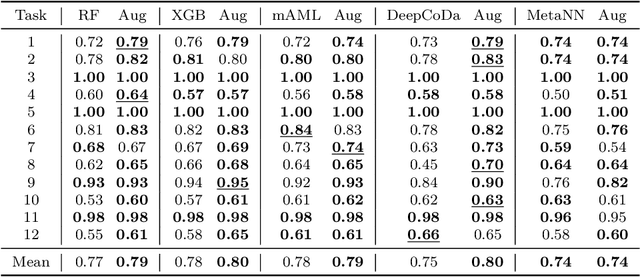
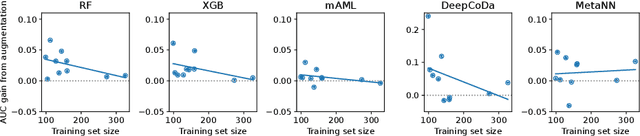
Abstract:Data augmentation plays a key role in modern machine learning pipelines. While numerous augmentation strategies have been studied in the context of computer vision and natural language processing, less is known for other data modalities. Our work extends the success of data augmentation to compositional data, i.e., simplex-valued data, which is of particular interest in the context of the human microbiome. Drawing on key principles from compositional data analysis, such as the Aitchison geometry of the simplex and subcompositions, we define novel augmentation strategies for this data modality. Incorporating our data augmentations into standard supervised learning pipelines results in consistent performance gains across a wide range of standard benchmark datasets. In particular, we set a new state-of-the-art for key disease prediction tasks including colorectal cancer, type 2 diabetes, and Crohn's disease. In addition, our data augmentations enable us to define a novel contrastive learning model, which improves on previous representation learning approaches for microbiome compositional data. Our code is available at https://github.com/cunningham-lab/AugCoDa.
The Three Ghosts of Medical AI: Can the Black-Box Present Deliver?
Dec 10, 2020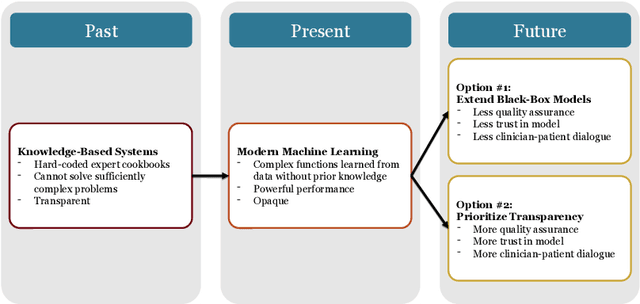
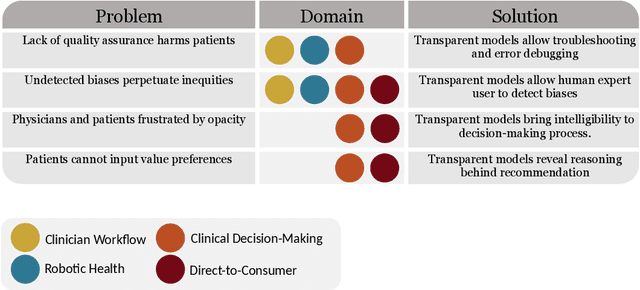
Abstract:Our title alludes to the three Christmas ghosts encountered by Ebenezer Scrooge in \textit{A Christmas Carol}, who guide Ebenezer through the past, present, and future of Christmas holiday events. Similarly, our article will take readers through a journey of the past, present, and future of medical AI. In doing so, we focus on the crux of modern machine learning: the reliance on powerful but intrinsically opaque models. When applied to the healthcare domain, these models fail to meet the needs for transparency that their clinician and patient end-users require. We review the implications of this failure, and argue that opaque models (1) lack quality assurance, (2) fail to elicit trust, and (3) restrict physician-patient dialogue. We then discuss how upholding transparency in all aspects of model design and model validation can help ensure the reliability of medical AI.
Trust and Medical AI: The challenges we face and the expertise needed to overcome them
Aug 18, 2020

Abstract:Artificial intelligence (AI) is increasingly of tremendous interest in the medical field. However, failures of medical AI could have serious consequences for both clinical outcomes and the patient experience. These consequences could erode public trust in AI, which could in turn undermine trust in our healthcare institutions. This article makes two contributions. First, it describes the major conceptual, technical, and humanistic challenges in medical AI. Second, it proposes a solution that hinges on the education and accreditation of new expert groups who specialize in the development, verification, and operation of medical AI technologies. These groups will be required to maintain trust in our healthcare institutions.
DeepCoDA: personalized interpretability for compositional health data
Jun 16, 2020



Abstract:Interpretability allows the domain-expert to directly evaluate the model's relevance and reliability, a practice that offers assurance and builds trust. In the healthcare setting, interpretable models should implicate relevant biological mechanisms independent of technical factors like data pre-processing. We define personalized interpretability as a measure of sample-specific feature attribution, and view it as a minimum requirement for a precision health model to justify its conclusions. Some health data, especially those generated by high-throughput sequencing experiments, have nuances that compromise precision health models and their interpretation. These data are compositional, meaning that each feature is conditionally dependent on all other features. We propose the Deep Compositional Data Analysis (DeepCoDA) framework to extend precision health modelling to high-dimensional compositional data, and to provide personalized interpretability through patient-specific weights. Our architecture maintains state-of-the-art performance across 25 real-world data sets, all while producing interpretations that are both personalized and fully coherent for compositional data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge