The Alzheimer's Disease Neuroimaging Initiative
ADNI
Transferring Models Trained on Natural Images to 3D MRI via Position Encoded Slice Models
Mar 02, 2023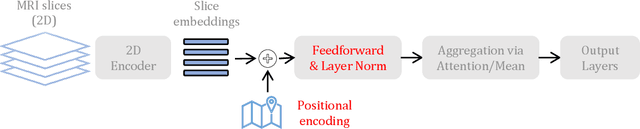
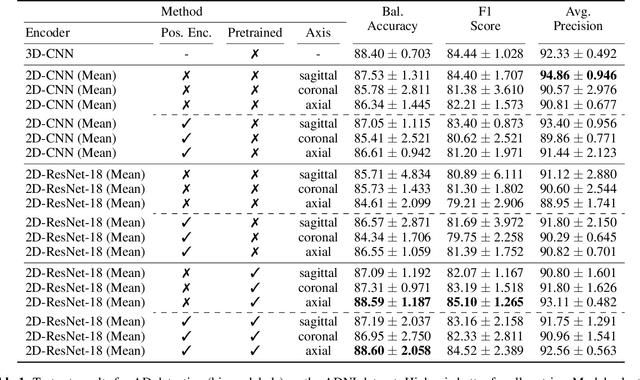
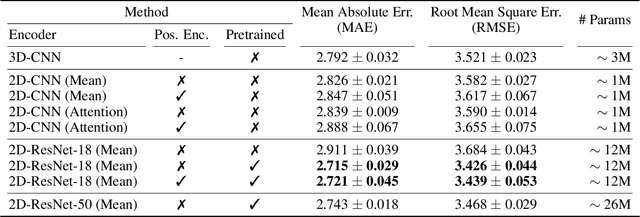
Abstract:Transfer learning has remarkably improved computer vision. These advances also promise improvements in neuroimaging, where training set sizes are often small. However, various difficulties arise in directly applying models pretrained on natural images to radiologic images, such as MRIs. In particular, a mismatch in the input space (2D images vs. 3D MRIs) restricts the direct transfer of models, often forcing us to consider only a few MRI slices as input. To this end, we leverage the 2D-Slice-CNN architecture of Gupta et al. (2021), which embeds all the MRI slices with 2D encoders (neural networks that take 2D image input) and combines them via permutation-invariant layers. With the insight that the pretrained model can serve as the 2D encoder, we initialize the 2D encoder with ImageNet pretrained weights that outperform those initialized and trained from scratch on two neuroimaging tasks -- brain age prediction on the UK Biobank dataset and Alzheimer's disease detection on the ADNI dataset. Further, we improve the modeling capabilities of 2D-Slice models by incorporating spatial information through position embeddings, which can improve the performance in some cases.
MRI-based Multi-task Decoupling Learning for Alzheimer's Disease Detection and MMSE Score Prediction: A Multi-site Validation
Apr 06, 2022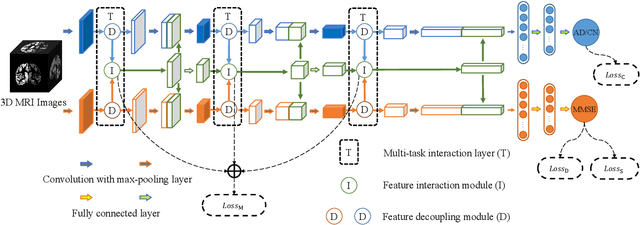

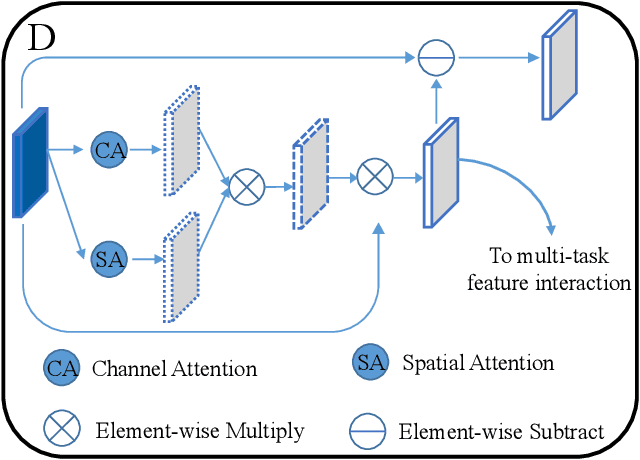

Abstract:Accurately detecting Alzheimer's disease (AD) and predicting mini-mental state examination (MMSE) score are important tasks in elderly health by magnetic resonance imaging (MRI). Most of the previous methods on these two tasks are based on single-task learning and rarely consider the correlation between them. Since the MMSE score, which is an important basis for AD diagnosis, can also reflect the progress of cognitive impairment, some studies have begun to apply multi-task learning methods to these two tasks. However, how to exploit feature correlation remains a challenging problem for these methods. To comprehensively address this challenge, we propose a MRI-based multi-task decoupled learning method for AD detection and MMSE score prediction. First, a multi-task learning network is proposed to implement AD detection and MMSE score prediction, which exploits feature correlation by adding three multi-task interaction layers between the backbones of the two tasks. Each multi-task interaction layer contains two feature decoupling modules and one feature interaction module. Furthermore, to enhance the generalization between tasks of the features selected by the feature decoupling module, we propose the feature consistency loss constrained feature decoupling module. Finally, in order to exploit the specific distribution information of MMSE score in different groups, a distribution loss is proposed to further enhance the model performance. We evaluate our proposed method on multi-site datasets. Experimental results show that our proposed multi-task decoupled representation learning method achieves good performance, outperforming single-task learning and other existing state-of-the-art methods.
High-dimensional regression over disease subgroups
Dec 09, 2016

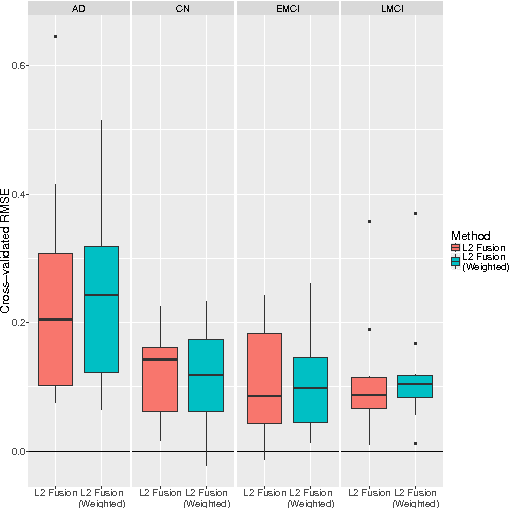
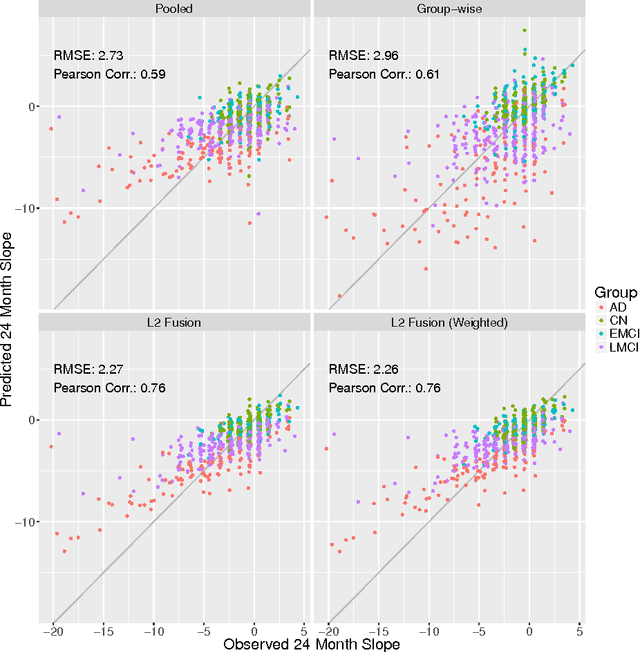
Abstract:We consider high-dimensional regression over subgroups of observations. Our work is motivated by biomedical problems, where disease subtypes, for example, may differ with respect to underlying regression models, but sample sizes at the subgroup-level may be limited. We focus on the case in which subgroup-specific models may be expected to be similar but not necessarily identical. Our approach is to treat subgroups as related problem instances and jointly estimate subgroup-specific regression coefficients. This is done in a penalized framework, combining an $\ell_1$ term with an additional term that penalizes differences between subgroup-specific coefficients. This gives solutions that are globally sparse but that allow information-sharing between the subgroups. We present algorithms for estimation and empirical results on simulated data and using Alzheimer's disease, amyotrophic lateral sclerosis and cancer datasets. These examples demonstrate the gains our approach can offer in terms of prediction and the ability to estimate subgroup-specific sparsity patterns.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge