Taufiq Hasan
Context-Aware Asymmetric Ensembling for Interpretable Retinopathy of Prematurity Screening via Active Query and Vascular Attention
Feb 05, 2026Abstract:Retinopathy of Prematurity (ROP) is among the major causes of preventable childhood blindness. Automated screening remains challenging, primarily due to limited data availability and the complex condition involving both structural staging and microvascular abnormalities. Current deep learning models depend heavily on large private datasets and passive multimodal fusion, which commonly fail to generalize on small, imbalanced public cohorts. We thus propose the Context-Aware Asymmetric Ensemble Model (CAA Ensemble) that simulates clinical reasoning through two specialized streams. First, the Multi-Scale Active Query Network (MS-AQNet) serves as a structure specialist, utilizing clinical contexts as dynamic query vectors to spatially control visual feature extraction for localization of the fibrovascular ridge. Secondly, VascuMIL encodes Vascular Topology Maps (VMAP) within a gated Multiple Instance Learning (MIL) network to precisely identify vascular tortuosity. A synergistic meta-learner ensembles these orthogonal signals to resolve diagnostic discordance across multiple objectives. Tested on a highly imbalanced cohort of 188 infants (6,004 images), the framework attained State-of-the-Art performance on two distinct clinical tasks: achieving a Macro F1-Score of 0.93 for Broad ROP staging and an AUC of 0.996 for Plus Disease detection. Crucially, the system features `Glass Box' transparency through counterfactual attention heatmaps and vascular threat maps, proving that clinical metadata dictates the model's visual search. Additionally, this study demonstrates that architectural inductive bias can serve as an effective bridge for the medical AI data gap.
Phi-SegNet: Phase-Integrated Supervision for Medical Image Segmentation
Jan 22, 2026Abstract:Deep learning has substantially advanced medical image segmentation, yet achieving robust generalization across diverse imaging modalities and anatomical structures remains a major challenge. A key contributor to this limitation lies in how existing architectures, ranging from CNNs to Transformers and their hybrids, primarily encode spatial information while overlooking frequency-domain representations that capture rich structural and textural cues. Although few recent studies have begun exploring spectral information at the feature level, supervision-level integration of frequency cues-crucial for fine-grained object localization-remains largely untapped. To this end, we propose Phi-SegNet, a CNN-based architecture that incorporates phase-aware information at both architectural and optimization levels. The network integrates Bi-Feature Mask Former (BFMF) modules that blend neighboring encoder features to reduce semantic gaps, and Reverse Fourier Attention (RFA) blocks that refine decoder outputs using phase-regularized features. A dedicated phase-aware loss aligns these features with structural priors, forming a closed feedback loop that emphasizes boundary precision. Evaluated on five public datasets spanning X-ray, US, histopathology, MRI, and colonoscopy, Phi-SegNet consistently achieved state-of-the-art performance, with an average relative improvement of 1.54+/-1.26% in IoU and 0.98+/-0.71% in F1-score over the next best-performing model. In cross-dataset generalization scenarios involving unseen datasets from the known domain, Phi-SegNet also exhibits robust and superior performance, highlighting its adaptability and modality-agnostic design. These findings demonstrate the potential of leveraging spectral priors in both feature representation and supervision, paving the way for generalized segmentation frameworks that excel in fine-grained object localization.
Weakly Supervised Tuberculosis Localization in Chest X-rays through Knowledge Distillation
Dec 11, 2025Abstract:Tuberculosis (TB) remains one of the leading causes of mortality worldwide, particularly in resource-limited countries. Chest X-ray (CXR) imaging serves as an accessible and cost-effective diagnostic tool but requires expert interpretation, which is often unavailable. Although machine learning models have shown high performance in TB classification, they often depend on spurious correlations and fail to generalize. Besides, building large datasets featuring high-quality annotations for medical images demands substantial resources and input from domain specialists, and typically involves several annotators reaching agreement, which results in enormous financial and logistical expenses. This study repurposes knowledge distillation technique to train CNN models reducing spurious correlations and localize TB-related abnormalities without requiring bounding-box annotations. By leveraging a teacher-student framework with ResNet50 architecture, the proposed method trained on TBX11k dataset achieve impressive 0.2428 mIOU score. Experimental results further reveal that the student model consistently outperforms the teacher, underscoring improved robustness and potential for broader clinical deployment in diverse settings.
A Multi-Stage Hybrid CNN-Transformer Network for Automated Pediatric Lung Sound Classification
Jul 27, 2025Abstract:Automated analysis of lung sound auscultation is essential for monitoring respiratory health, especially in regions facing a shortage of skilled healthcare workers. While respiratory sound classification has been widely studied in adults, its ap plication in pediatric populations, particularly in children aged <6 years, remains an underexplored area. The developmental changes in pediatric lungs considerably alter the acoustic proper ties of respiratory sounds, necessitating specialized classification approaches tailored to this age group. To address this, we propose a multistage hybrid CNN-Transformer framework that combines CNN-extracted features with an attention-based architecture to classify pediatric respiratory diseases using scalogram images from both full recordings and individual breath events. Our model achieved an overall score of 0.9039 in binary event classifi cation and 0.8448 in multiclass event classification by employing class-wise focal loss to address data imbalance. At the recording level, the model attained scores of 0.720 for ternary and 0.571 for multiclass classification. These scores outperform the previous best models by 3.81% and 5.94%, respectively. This approach offers a promising solution for scalable pediatric respiratory disease diagnosis, especially in resource-limited settings.
LMLCC-Net: A Semi-Supervised Deep Learning Model for Lung Nodule Malignancy Prediction from CT Scans using a Novel Hounsfield Unit-Based Intensity Filtering
May 09, 2025Abstract:Lung cancer is the leading cause of patient mortality in the world. Early diagnosis of malignant pulmonary nodules in CT images can have a significant impact on reducing disease mortality and morbidity. In this work, we propose LMLCC-Net, a novel deep learning framework for classifying nodules from CT scan images using a 3D CNN, considering Hounsfield Unit (HU)-based intensity filtering. Benign and malignant nodules have significant differences in their intensity profile of HU, which was not exploited in the literature. Our method considers the intensity pattern as well as the texture for the prediction of malignancies. LMLCC-Net extracts features from multiple branches that each use a separate learnable HU-based intensity filtering stage. Various combinations of branches and learnable ranges of filters were explored to finally produce the best-performing model. In addition, we propose a semi-supervised learning scheme for labeling ambiguous cases and also developed a lightweight model to classify the nodules. The experimental evaluations are carried out on the LUNA16 dataset. Our proposed method achieves a classification accuracy (ACC) of 91.96%, a sensitivity (SEN) of 92.04%, and an area under the curve (AUC) of 91.87%, showing improved performance compared to existing methods. The proposed method can have a significant impact in helping radiologists in the classification of pulmonary nodules and improving patient care.
Domain Generalization for Improved Human Activity Recognition in Office Space Videos Using Adaptive Pre-processing
Mar 16, 2025
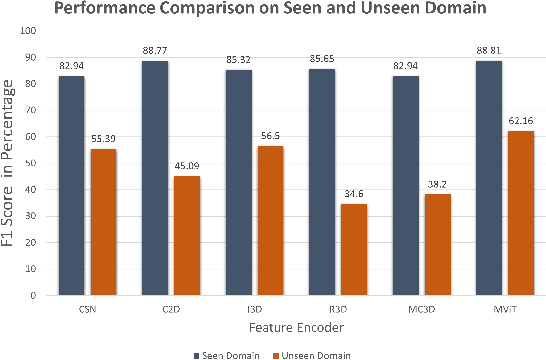
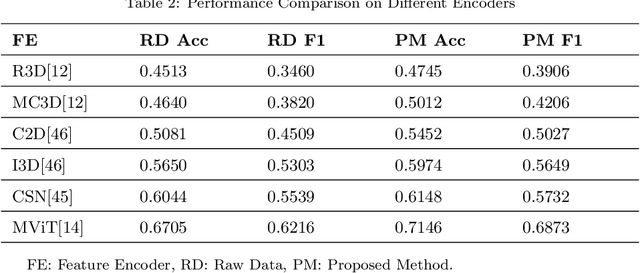
Abstract:Automatic video activity recognition is crucial across numerous domains like surveillance, healthcare, and robotics. However, recognizing human activities from video data becomes challenging when training and test data stem from diverse domains. Domain generalization, adapting to unforeseen domains, is thus essential. This paper focuses on office activity recognition amidst environmental variability. We propose three pre-processing techniques applicable to any video encoder, enhancing robustness against environmental variations. Our study showcases the efficacy of MViT, a leading state-of-the-art video classification model, and other video encoders combined with our techniques, outperforming state-of-the-art domain adaptation methods. Our approach significantly boosts accuracy, precision, recall and F1 score on unseen domains, emphasizing its adaptability in real-world scenarios with diverse video data sources. This method lays a foundation for more reliable video activity recognition systems across heterogeneous data domains.
BUET Multi-disease Heart Sound Dataset: A Comprehensive Auscultation Dataset for Developing Computer-Aided Diagnostic Systems
Sep 01, 2024Abstract:Cardiac auscultation, an integral tool in diagnosing cardiovascular diseases (CVDs), often relies on the subjective interpretation of clinicians, presenting a limitation in consistency and accuracy. Addressing this, we introduce the BUET Multi-disease Heart Sound (BMD-HS) dataset - a comprehensive and meticulously curated collection of heart sound recordings. This dataset, encompassing 864 recordings across five distinct classes of common heart sounds, represents a broad spectrum of valvular heart diseases, with a focus on diagnostically challenging cases. The standout feature of the BMD-HS dataset is its innovative multi-label annotation system, which captures a diverse range of diseases and unique disease states. This system significantly enhances the dataset's utility for developing advanced machine learning models in automated heart sound classification and diagnosis. By bridging the gap between traditional auscultation practices and contemporary data-driven diagnostic methods, the BMD-HS dataset is poised to revolutionize CVD diagnosis and management, providing an invaluable resource for the advancement of cardiac health research. The dataset is publicly available at this link: https://github.com/mHealthBuet/BMD-HS-Dataset.
Improving Pediatric Pneumonia Diagnosis with Adult Chest X-ray Images Utilizing Contrastive Learning and Embedding Similarity
Apr 19, 2024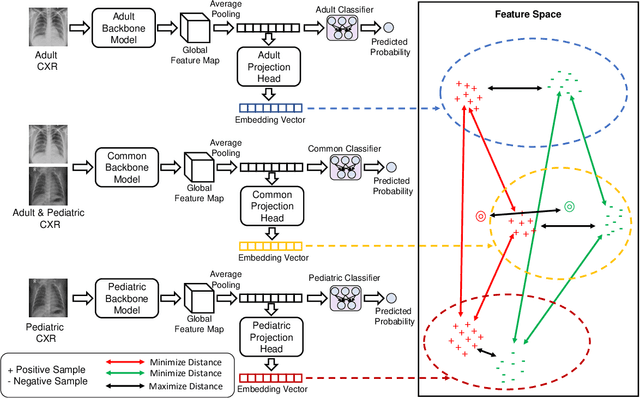
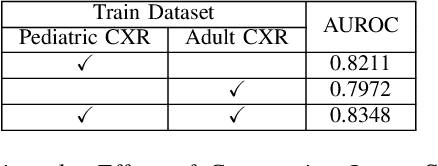
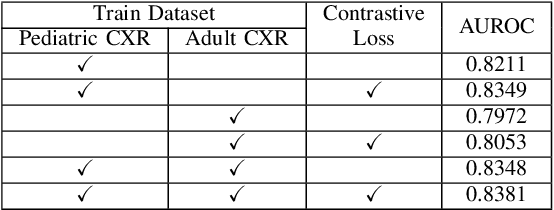

Abstract:Despite the advancement of deep learning-based computer-aided diagnosis (CAD) methods for pneumonia from adult chest x-ray (CXR) images, the performance of CAD methods applied to pediatric images remains suboptimal, mainly due to the lack of large-scale annotated pediatric imaging datasets. Establishing a proper framework to leverage existing adult large-scale CXR datasets can thus enhance pediatric pneumonia detection performance. In this paper, we propose a three-branch parallel path learning-based framework that utilizes both adult and pediatric datasets to improve the performance of deep learning models on pediatric test datasets. The paths are trained with pediatric only, adult only, and both types of CXRs, respectively. Our proposed framework utilizes the multi-positive contrastive loss to cluster the classwise embeddings and the embedding similarity loss among these three parallel paths to make the classwise embeddings as close as possible to reduce the effect of domain shift. Experimental evaluations on open-access adult and pediatric CXR datasets show that the proposed method achieves a superior AUROC score of 0.8464 compared to 0.8348 obtained using the conventional approach of join training on both datasets. The proposed approach thus paves the way for generalized CAD models that are effective for both adult and pediatric age groups.
Uformer: A UNet-Transformer fused robust end-to-end deep learning framework for real-time denoising of lung sounds
Apr 05, 2024Abstract:Objective: Lung auscultation is a valuable tool in diagnosing and monitoring various respiratory diseases. However, lung sounds (LS) are significantly affected by numerous sources of contamination, especially when recorded in real-world clinical settings. Conventional denoising models prove impractical for LS denoising, primarily owing to spectral overlap complexities arising from diverse noise sources. To address this issue, we propose a specialized deep-learning model (Uformer) for lung sound denoising. Methods: The proposed Uformer model is constituted of three modules: a Convolutional Neural Network (CNN) encoder module, dedicated to extracting latent features; a Transformer encoder module, employed to further enhance the encoding of unique LS features and effectively capture intricate long-range dependencies; and a CNN decoder module, employed to generate the denoised signals. An ablation study was performed in order to find the most optimal architecture. Results: The performance of the proposed Uformer model was evaluated on lung sounds induced with different types of synthetic and real-world noises. Lung sound signals of -12 dB to 15 dB signal-to-noise ratio (SNR) were considered in testing experiments. The proposed model showed an average SNR improvement of 16.51 dB when evaluated with -12 dB LS signals. Our end-to-end model, with an average SNR improvement of 19.31 dB, outperforms the existing model when evaluated with ambient noise and fewer parameters. Conclusion: Based on the qualitative and quantitative findings in this study, it can be stated that Uformer is robust and generalized to be used in assisting the monitoring of respiratory conditions.
A Web-based Mpox Skin Lesion Detection System Using State-of-the-art Deep Learning Models Considering Racial Diversity
Jun 25, 2023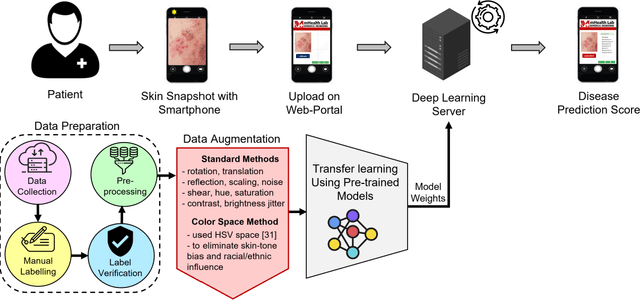
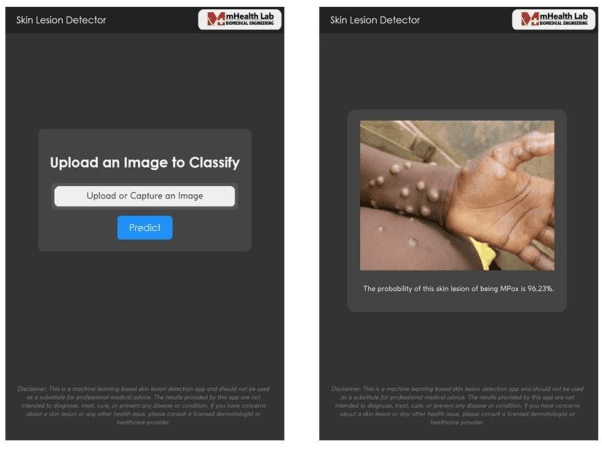
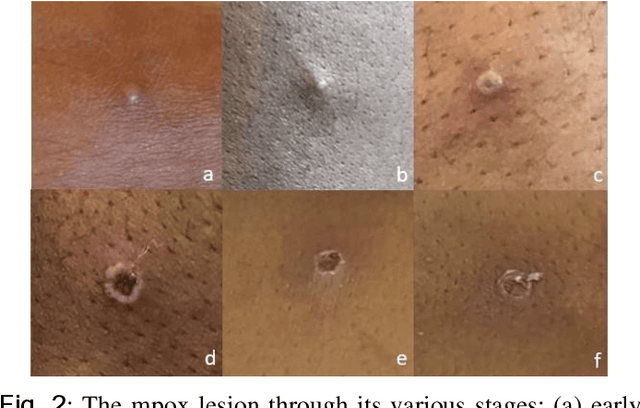
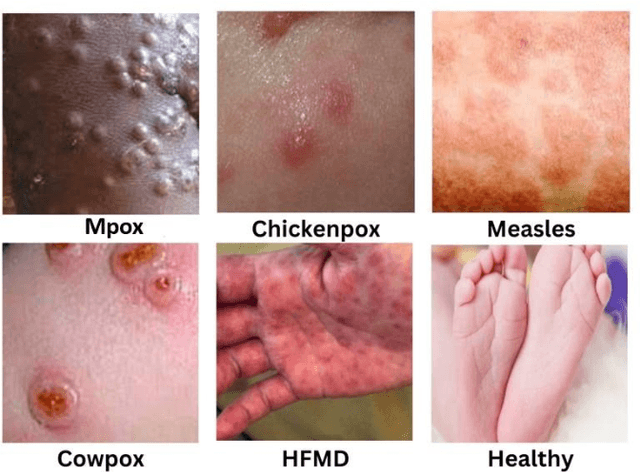
Abstract:The recent 'Mpox' outbreak, formerly known as 'Monkeypox', has become a significant public health concern and has spread to over 110 countries globally. The challenge of clinically diagnosing mpox early on is due, in part, to its similarity to other types of rashes. Computer-aided screening tools have been proven valuable in cases where Polymerase Chain Reaction (PCR) based diagnosis is not immediately available. Deep learning methods are powerful in learning complex data representations, but their efficacy largely depends on adequate training data. To address this challenge, we present the "Mpox Skin Lesion Dataset Version 2.0 (MSLD v2.0)" as a follow-up to the previously released openly accessible dataset, one of the first datasets containing mpox lesion images. This dataset contains images of patients with mpox and five other non-mpox classes (chickenpox, measles, hand-foot-mouth disease, cowpox, and healthy). We benchmark the performance of several state-of-the-art deep learning models, including VGG16, ResNet50, DenseNet121, MobileNetV2, EfficientNetB3, InceptionV3, and Xception, to classify mpox and other infectious skin diseases. In order to reduce the impact of racial bias, we utilize a color space data augmentation method to increase skin color variability during training. Additionally, by leveraging transfer learning implemented with pre-trained weights generated from the HAM10000 dataset, an extensive collection of pigmented skin lesion images, we achieved the best overall accuracy of $83.59\pm2.11\%$. Finally, the developed models are incorporated within a prototype web application to analyze uploaded skin images by a user and determine whether a subject is a suspected mpox patient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge