Md. Aynal Haque
ResDTA: Predicting Drug-Target Binding Affinity Using Residual Skip Connections
Mar 20, 2023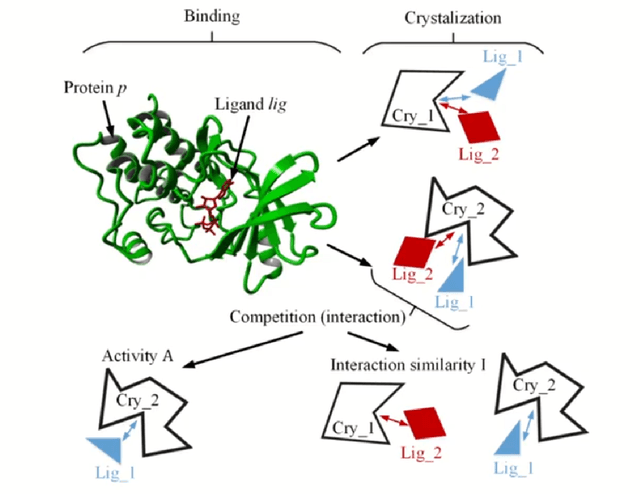
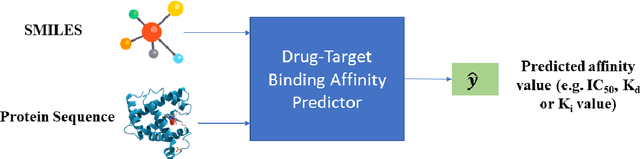
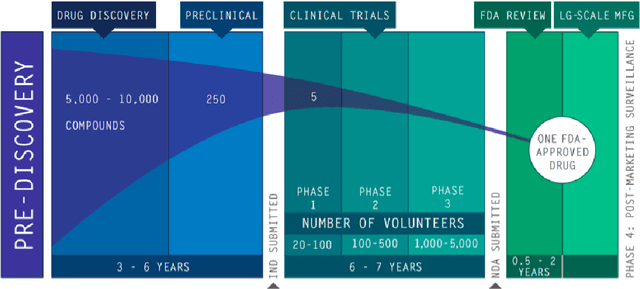
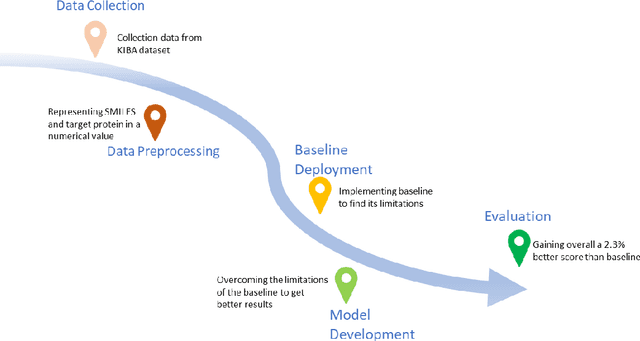
Abstract:The discovery of novel drug target (DT) interactions is an important step in the drug development process. The majority of computer techniques for predicting DT interactions have focused on binary classification, with the goal of determining whether or not a DT pair interacts. Protein ligand interactions, on the other hand, assume a continuous range of binding strength values, also known as binding affinity, and forecasting this value remains a difficulty. As the amount of affinity data in DT knowledge-bases grows, advanced learning techniques such as deep learning architectures can be used to predict binding affinities. In this paper, we present a deep-learning-based methodology for predicting DT binding affinities using just sequencing information from both targets and drugs. The results show that the proposed deep learning-based model that uses the 1D representations of targets and drugs is an effective approach for drug target binding affinity prediction and it does not require additional chemical domain knowledge to work with. The model in which high-level representations of a drug and a target are constructed via CNNs that uses residual skip connections and also with an additional stream to create a high-level combined representation of the drug-target pair achieved the best Concordance Index (CI) performance in one of the largest benchmark datasets, outperforming the recent state-of-the-art method AttentionDTA and many other machine-learning and deep-learning based baseline methods for DT binding affinity prediction that uses the 1D representations of targets and drugs.
Learning to Generalize towards Unseen Domains via a Content-Aware Style Invariant Framework for Disease Detection from Chest X-rays
Feb 27, 2023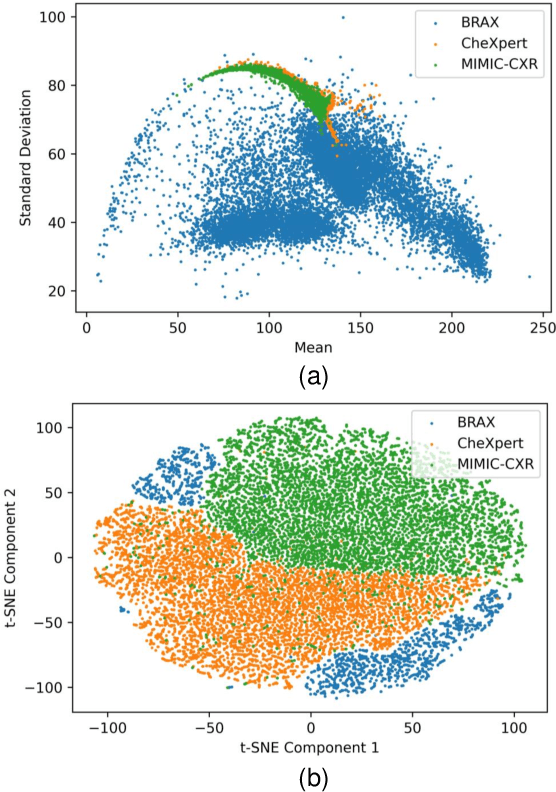
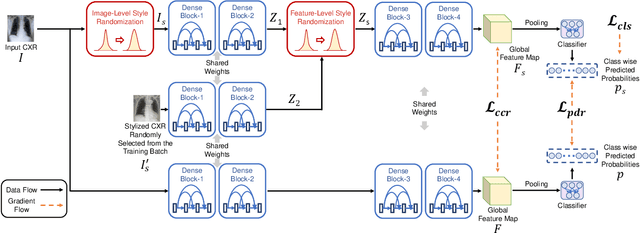
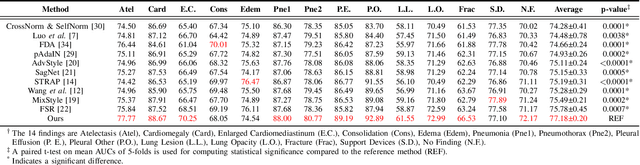
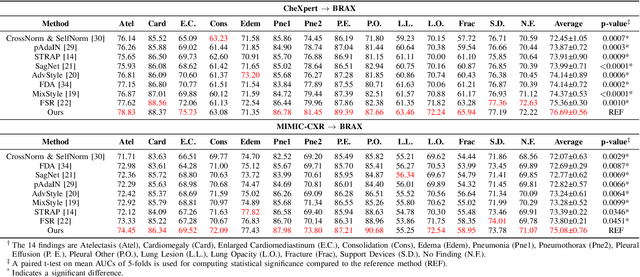
Abstract:Performance degradation due to source domain mismatch is a longstanding challenge in deep learning-based medical image analysis, particularly for chest X-rays. Several methods have been proposed to address this domain shift, such as utilizing adversarial learning or multi-domain mixups to extract domain-invariant high-level features. However, these methods do not explicitly account for or regularize the content and style attributes of the extracted domain-invariant features. Recent studies have demonstrated that CNN models exhibit a strong bias toward styles (i.e., textures) rather than content, in stark contrast to the human-vision system. Explainable representations are paramount for a robust and generalizable understanding of medical images. Thus, the learned high-level semantic features need to be both content-specific, i.e., pathology-specific and domain-agnostic, as well as style invariant. Inspired by this, we propose a novel framework that improves cross-domain performances by focusing more on content while reducing style bias. We employ a style randomization module at both image and feature levels to create stylized perturbation features while preserving the content using an end-to-end framework. We extract the global features from the backbone model for the same chest X-ray with and without style randomized. We apply content consistency regularization between them to tweak the framework's sensitivity toward content markers for accurate predictions. Extensive experiments on unseen domain test datasets demonstrate that our proposed pipeline is more robust in the presence of domain shifts and achieves state-of-the-art performance. Our code is available via https://github.com/rafizunaed/domain_agnostic_content_aware_style_invariant.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge