Mohammad Zunaed
Real Time NILM Based Power Monitoring of Identical Induction Motors Representing Cutting Machines in Textile Industry
Jan 04, 2026Abstract:The textile industry in Bangladesh is one of the most energy-intensive sectors, yet its monitoring practices remain largely outdated, resulting in inefficient power usage and high operational costs. To address this, we propose a real-time Non-Intrusive Load Monitoring (NILM)-based framework tailored for industrial applications, with a focus on identical motor-driven loads representing textile cutting machines. A hardware setup comprising voltage and current sensors, Arduino Mega and ESP8266 was developed to capture aggregate and individual load data, which was stored and processed on cloud platforms. A new dataset was created from three identical induction motors and auxiliary loads, totaling over 180,000 samples, to evaluate the state-of-the-art MATNILM model under challenging industrial conditions. Results indicate that while aggregate energy estimation was reasonably accurate, per-appliance disaggregation faced difficulties, particularly when multiple identical machines operated simultaneously. Despite these challenges, the integrated system demonstrated practical real-time monitoring with remote accessibility through the Blynk application. This work highlights both the potential and limitations of NILM in industrial contexts, offering insights into future improvements such as higher-frequency data collection, larger-scale datasets and advanced deep learning approaches for handling identical loads.
Domain Generalization for Improved Human Activity Recognition in Office Space Videos Using Adaptive Pre-processing
Mar 16, 2025
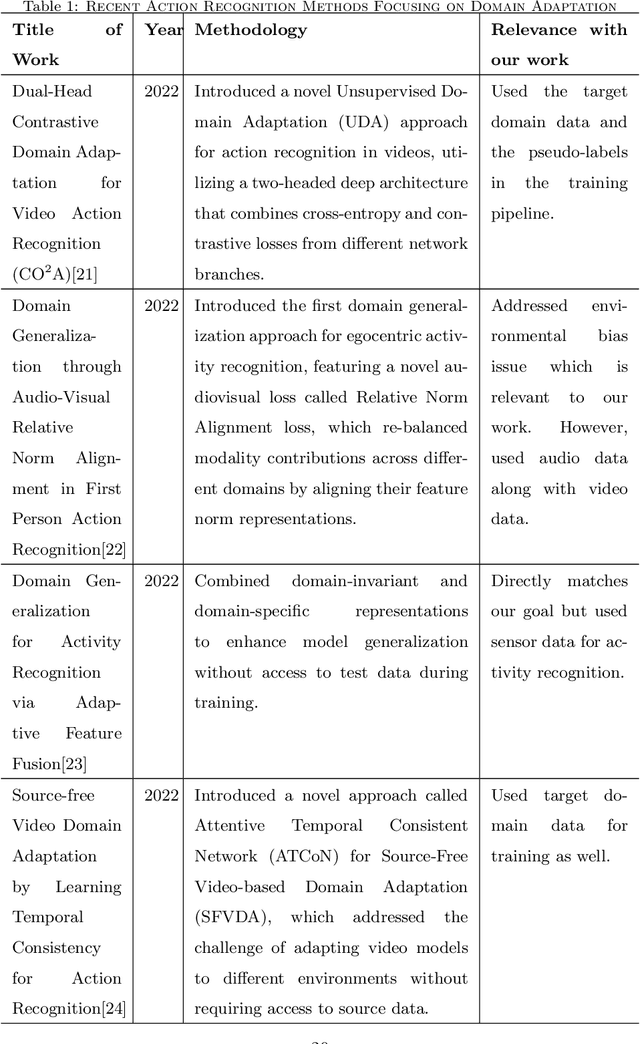
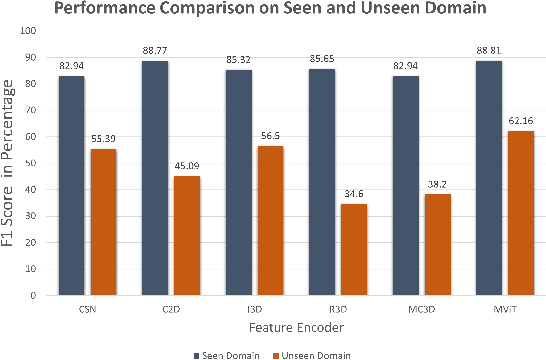
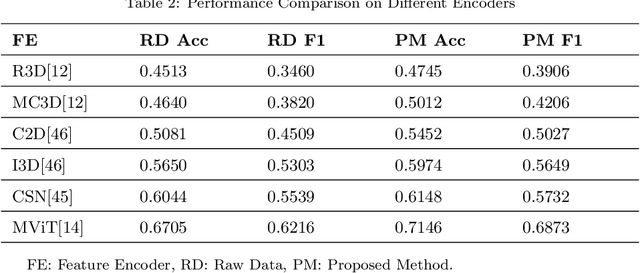
Abstract:Automatic video activity recognition is crucial across numerous domains like surveillance, healthcare, and robotics. However, recognizing human activities from video data becomes challenging when training and test data stem from diverse domains. Domain generalization, adapting to unforeseen domains, is thus essential. This paper focuses on office activity recognition amidst environmental variability. We propose three pre-processing techniques applicable to any video encoder, enhancing robustness against environmental variations. Our study showcases the efficacy of MViT, a leading state-of-the-art video classification model, and other video encoders combined with our techniques, outperforming state-of-the-art domain adaptation methods. Our approach significantly boosts accuracy, precision, recall and F1 score on unseen domains, emphasizing its adaptability in real-world scenarios with diverse video data sources. This method lays a foundation for more reliable video activity recognition systems across heterogeneous data domains.
Improving Pediatric Pneumonia Diagnosis with Adult Chest X-ray Images Utilizing Contrastive Learning and Embedding Similarity
Apr 19, 2024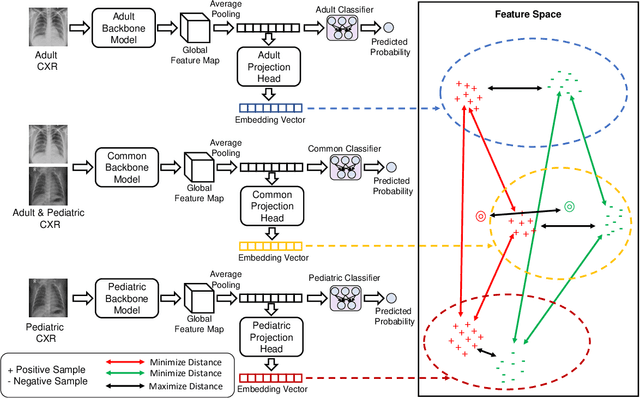
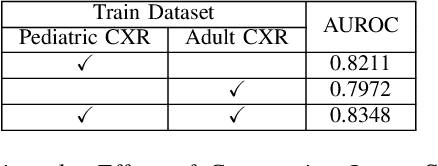
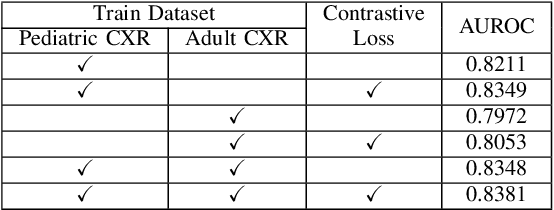

Abstract:Despite the advancement of deep learning-based computer-aided diagnosis (CAD) methods for pneumonia from adult chest x-ray (CXR) images, the performance of CAD methods applied to pediatric images remains suboptimal, mainly due to the lack of large-scale annotated pediatric imaging datasets. Establishing a proper framework to leverage existing adult large-scale CXR datasets can thus enhance pediatric pneumonia detection performance. In this paper, we propose a three-branch parallel path learning-based framework that utilizes both adult and pediatric datasets to improve the performance of deep learning models on pediatric test datasets. The paths are trained with pediatric only, adult only, and both types of CXRs, respectively. Our proposed framework utilizes the multi-positive contrastive loss to cluster the classwise embeddings and the embedding similarity loss among these three parallel paths to make the classwise embeddings as close as possible to reduce the effect of domain shift. Experimental evaluations on open-access adult and pediatric CXR datasets show that the proposed method achieves a superior AUROC score of 0.8464 compared to 0.8348 obtained using the conventional approach of join training on both datasets. The proposed approach thus paves the way for generalized CAD models that are effective for both adult and pediatric age groups.
Learning to Generalize towards Unseen Domains via a Content-Aware Style Invariant Framework for Disease Detection from Chest X-rays
Feb 27, 2023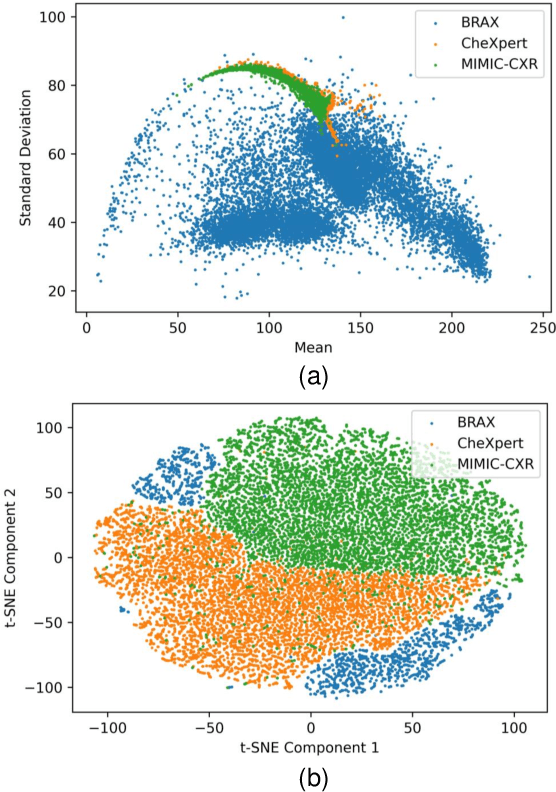
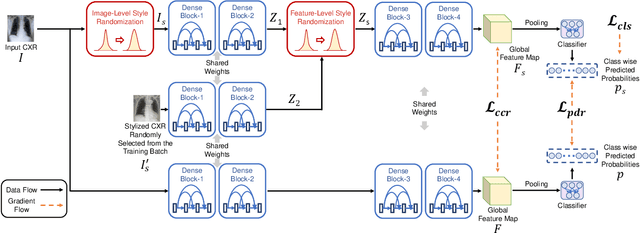
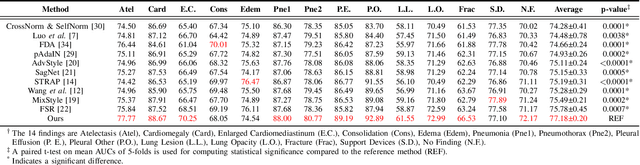
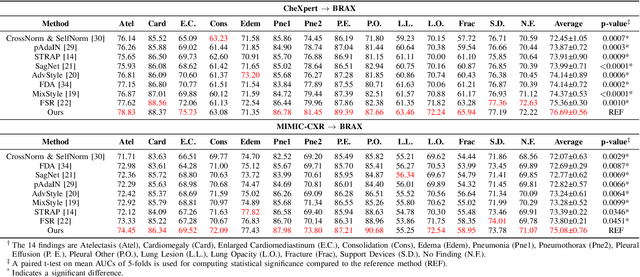
Abstract:Performance degradation due to source domain mismatch is a longstanding challenge in deep learning-based medical image analysis, particularly for chest X-rays. Several methods have been proposed to address this domain shift, such as utilizing adversarial learning or multi-domain mixups to extract domain-invariant high-level features. However, these methods do not explicitly account for or regularize the content and style attributes of the extracted domain-invariant features. Recent studies have demonstrated that CNN models exhibit a strong bias toward styles (i.e., textures) rather than content, in stark contrast to the human-vision system. Explainable representations are paramount for a robust and generalizable understanding of medical images. Thus, the learned high-level semantic features need to be both content-specific, i.e., pathology-specific and domain-agnostic, as well as style invariant. Inspired by this, we propose a novel framework that improves cross-domain performances by focusing more on content while reducing style bias. We employ a style randomization module at both image and feature levels to create stylized perturbation features while preserving the content using an end-to-end framework. We extract the global features from the backbone model for the same chest X-ray with and without style randomized. We apply content consistency regularization between them to tweak the framework's sensitivity toward content markers for accurate predictions. Extensive experiments on unseen domain test datasets demonstrate that our proposed pipeline is more robust in the presence of domain shifts and achieves state-of-the-art performance. Our code is available via https://github.com/rafizunaed/domain_agnostic_content_aware_style_invariant.
A Novel Hierarchical-Classification-Block Based Convolutional Neural Network for Source Camera Model Identification
Dec 08, 2022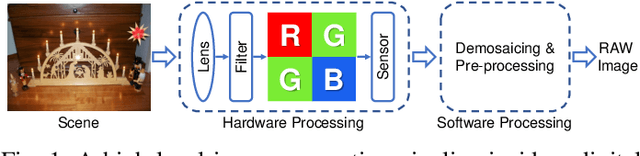
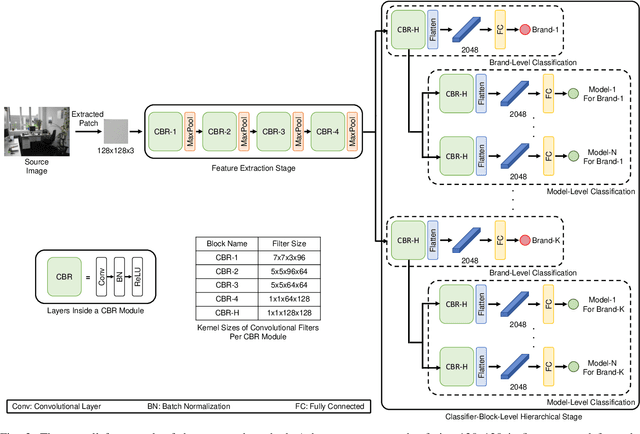


Abstract:Digital security has been an active area of research interest due to the rapid adaptation of internet infrastructure, the increasing popularity of social media, and digital cameras. Due to inherent differences in working principles to generate an image, different camera brands left behind different intrinsic processing noises which can be used to identify the camera brand. In the last decade, many signal processing and deep learning-based methods have been proposed to identify and isolate this noise from the scene details in an image to detect the source camera brand. One prominent solution is to utilize a hierarchical classification system rather than the traditional single-classifier approach. Different individual networks are used for brand-level and model-level source camera identification. This approach allows for better scaling and requires minimal modifications for adding a new camera brand/model to the solution. However, using different full-fledged networks for both brand and model-level classification substantially increases memory consumption and training complexity. Moreover, extracted low-level features from the different network's initial layers often coincide, resulting in redundant weights. To mitigate the training and memory complexity, we propose a classifier-block-level hierarchical system instead of a network-level one for source camera model classification. Our proposed approach not only results in significantly fewer parameters but also retains the capability to add a new camera model with minimal modification. Thorough experimentation on the publicly available Dresden dataset shows that our proposed approach can achieve the same level of state-of-the-art performance but requires fewer parameters compared to a state-of-the-art network-level hierarchical-based system.
A Novel Attention Mechanism Using Anatomical Prior Probability Maps for Thoracic Disease Classification from X-Ray Images
Oct 06, 2022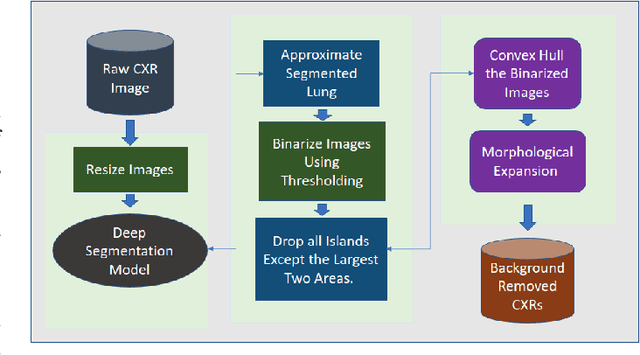
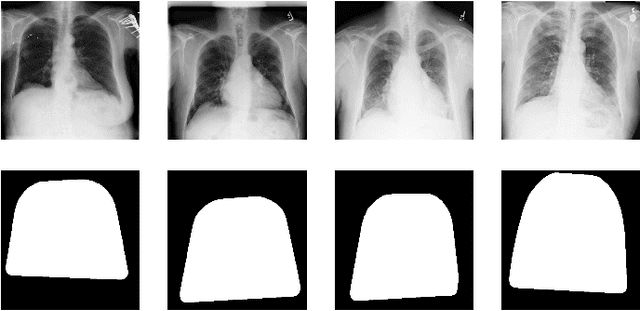
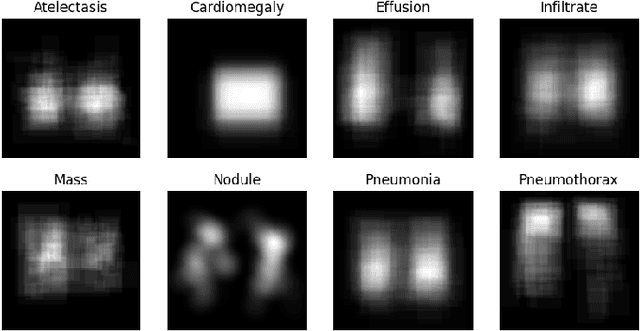
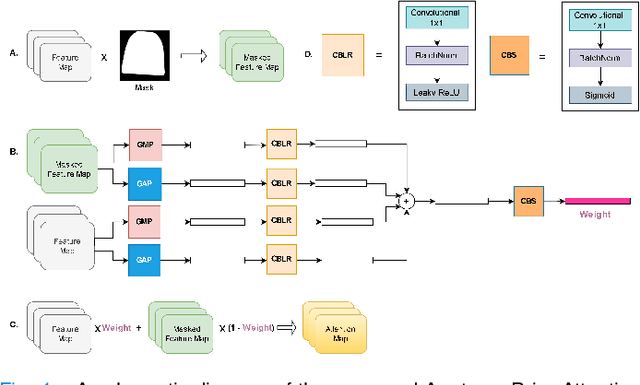
Abstract:Computer-aided disease diagnosis and prognosis based on medical images is a rapidly emerging field. Many Convolutional Neural Network (CNN) architectures have been developed by researchers for disease classification and localization from chest X-ray images. It is known that different thoracic disease lesions are more likely to occur in specific anatomical regions compared to others. Based on this knowledge, we first estimate a disease-dependent spatial probability, i.e., an anatomical prior, that indicates the probability of occurrence of a disease in a specific region in a chest X-ray image. Next, we develop a novel attention-based classification model that combines information from the estimated anatomical prior and automatically extracted chest region of interest (ROI) masks to provide attention to the feature maps generated from a deep convolution network. Unlike previous works that utilize various self-attention mechanisms, the proposed method leverages the extracted chest ROI masks along with the probabilistic anatomical prior information, which selects the region of interest for different diseases to provide attention. The proposed method shows superior performance in disease classification on the NIH ChestX-ray14 dataset compared to existing state-of-the-art methods while reaching an area under the ROC curve (AUC) of 0.8427. Regarding disease localization, the proposed method shows competitive performance compared to state-of-the-art methods, achieving an accuracy of 61% with an Intersection over Union (IoU) threshold of 0.3. The proposed method can also be generalized to other medical image-based disease classification and localization tasks where the probability of occurrence of the lesion is dependent on specific anatomical sites.
Anatomy X-Net: A Semi-Supervised Anatomy Aware Convolutional Neural Network for Thoracic Disease Classification
Jun 10, 2021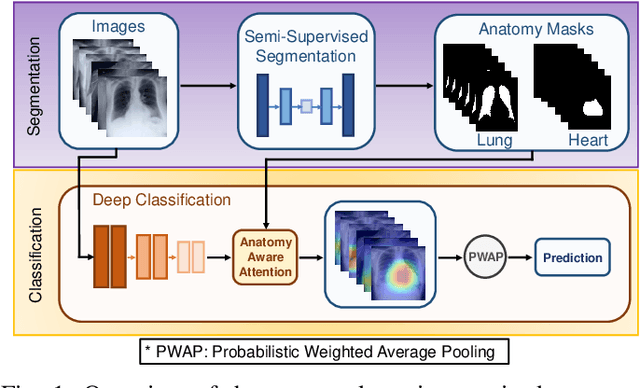
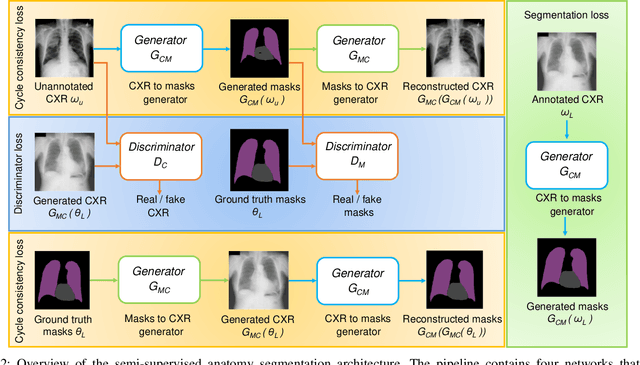
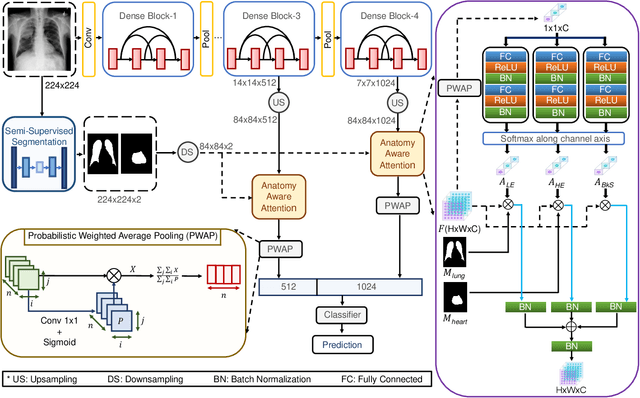
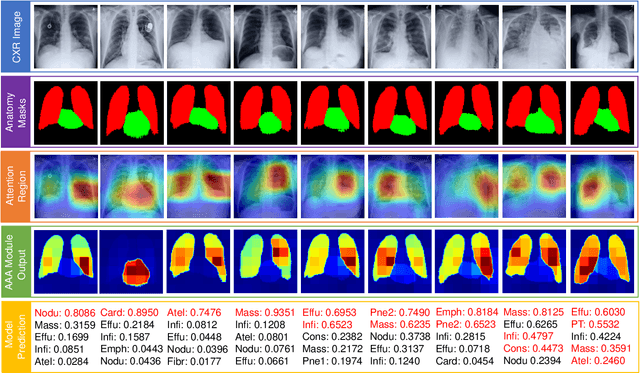
Abstract:Thoracic disease detection from chest radiographs using deep learning methods has been an active area of research in the last decade. Most previous methods attempt to focus on the diseased organs of the image by identifying spatial regions responsible for significant contributions to the model's prediction. In contrast, expert radiologists first locate the prominent anatomical structures before determining if those regions are anomalous. Therefore, integrating anatomical knowledge within deep learning models could bring substantial improvement in automatic disease classification. This work proposes an anatomy-aware attention-based architecture named Anatomy X-Net, that prioritizes the spatial features guided by the pre-identified anatomy regions. We leverage a semi-supervised learning method using the JSRT dataset containing organ-level annotation to obtain the anatomical segmentation masks (for lungs and heart) for the NIH and CheXpert datasets. The proposed Anatomy X-Net uses the pre-trained DenseNet-121 as the backbone network with two corresponding structured modules, the Anatomy Aware Attention (AAA) and Probabilistic Weighted Average Pooling (PWAP), in a cohesive framework for anatomical attention learning. Our proposed method sets new state-of-the-art performance on the official NIH test set with an AUC score of 0.8439, proving the efficacy of utilizing the anatomy segmentation knowledge to improve the thoracic disease classification. Furthermore, the Anatomy X-Net yields an averaged AUC of 0.9020 on the Stanford CheXpert dataset, improving on existing methods that demonstrate the generalizability of the proposed framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge