Suman Sedai
Uncertainty guided semi-supervised segmentation of retinal layers in OCT images
Mar 02, 2021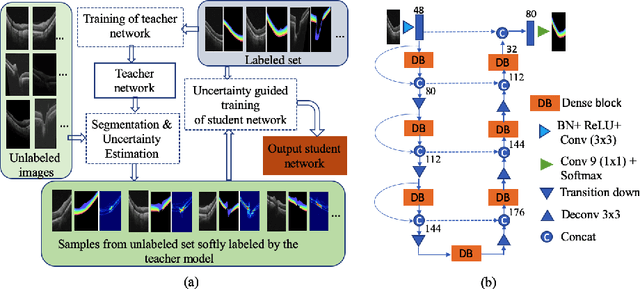
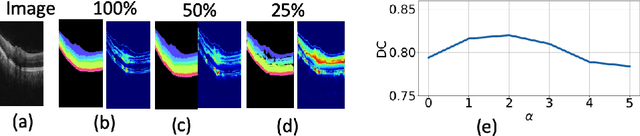
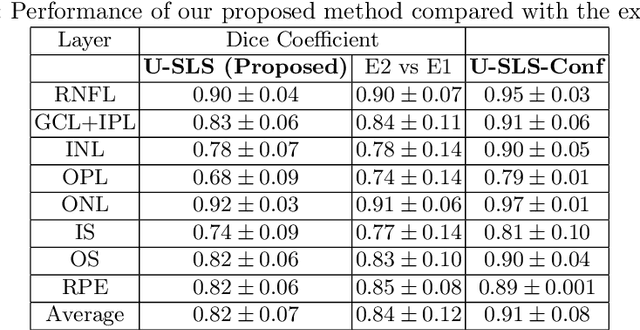
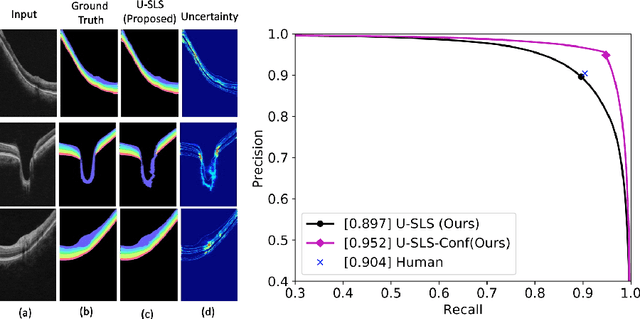
Abstract:Deep convolutional neural networks have shown outstanding performance in medical image segmentation tasks. The usual problem when training supervised deep learning methods is the lack of labeled data which is time-consuming and costly to obtain. In this paper, we propose a novel uncertainty-guided semi-supervised learning based on a student-teacher approach for training the segmentation network using limited labeled samples and a large number of unlabeled images. First, a teacher segmentation model is trained from the labeled samples using Bayesian deep learning. The trained model is used to generate soft segmentation labels and uncertainty maps for the unlabeled set. The student model is then updated using the softly segmented samples and the corresponding pixel-wise confidence of the segmentation quality estimated from the uncertainty of the teacher model using a newly designed loss function. Experimental results on a retinal layer segmentation task show that the proposed method improves the segmentation performance in comparison to the fully supervised approach and is on par with the expert annotator. The proposed semi-supervised segmentation framework is a key contribution and applicable for biomedical image segmentation across various imaging modalities where access to annotated medical images is challenging
* MICCAI,19
Analyzing Epistemic and Aleatoric Uncertainty for Drusen Segmentation in Optical Coherence Tomography Images
Feb 08, 2021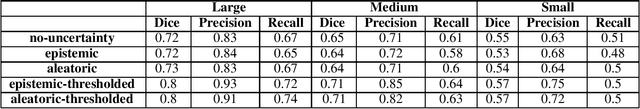
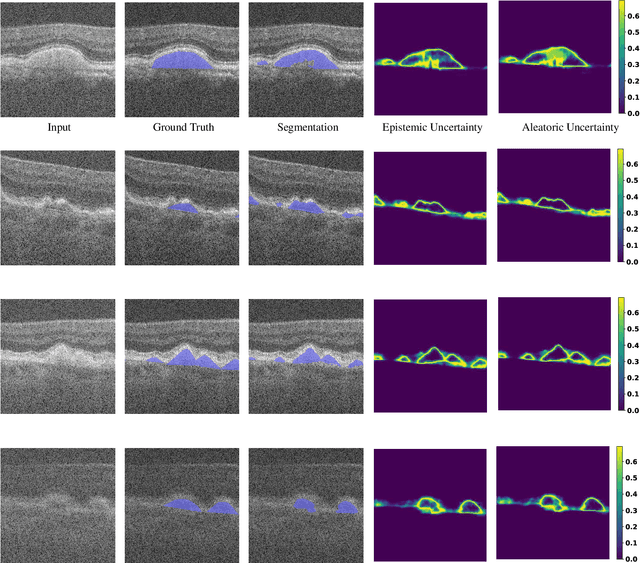
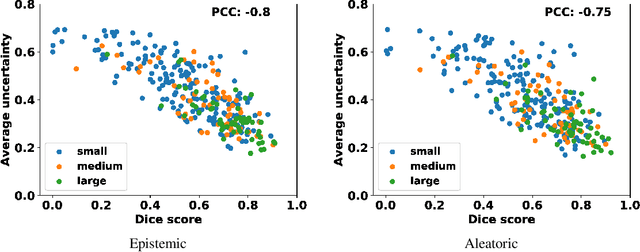
Abstract:Age-related macular degeneration (AMD) is one of the leading causes of permanent vision loss in people aged over 60 years. Accurate segmentation of biomarkers such as drusen that points to the early stages of AMD is crucial in preventing further vision impairment. However, segmenting drusen is extremely challenging due to their varied sizes and appearances, low contrast and noise resemblance. Most existing literature, therefore, have focused on size estimation of drusen using classification, leaving the challenge of accurate segmentation less tackled. Additionally, obtaining the pixel-wise annotations is extremely costly and such labels can often be noisy, suffering from inter-observer and intra-observer variability. Quantification of uncertainty associated with segmentation tasks offers principled measures to inspect the segmentation output. Realizing its utility in identifying erroneous segmentation and the potential applications in clinical decision making, here we develop a U-Net based drusen segmentation model and quantify the segmentation uncertainty. We investigate epistemic and aleatoric uncertainty capturing model confidence and data uncertainty respectively. We present segmentation results and show how uncertainty can help formulate robust evaluation strategies. We visually inspect the pixel-wise uncertainty and segmentation results on test images. We finally analyze the correlation between segmentation uncertainty and accuracy. Our results demonstrate the utility of leveraging uncertainties in developing and explaining segmentation models for medical image analysis.
Dueling Deep Q-Network for Unsupervised Inter-frame Eye Movement Correction in Optical Coherence Tomography Volumes
Jul 03, 2020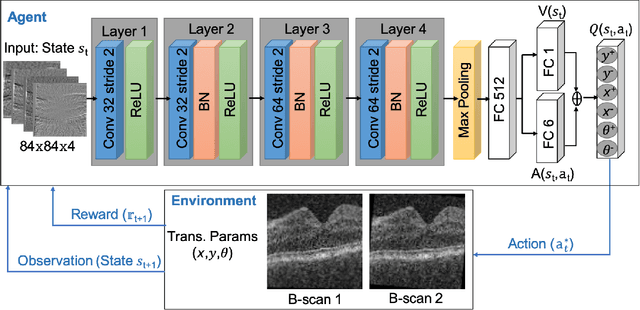
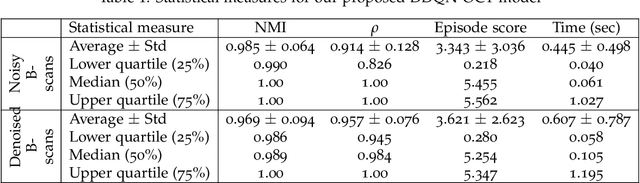
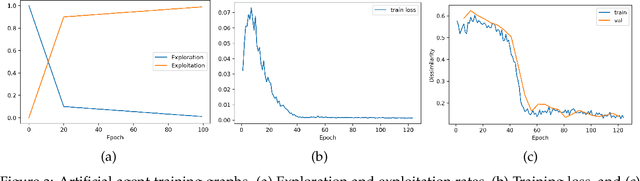
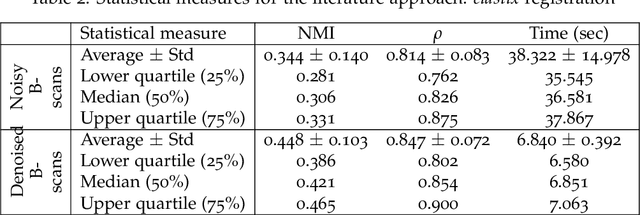
Abstract:In optical coherence tomography (OCT) volumes of retina, the sequential acquisition of the individual slices makes this modality prone to motion artifacts, misalignments between adjacent slices being the most noticeable. Any distortion in OCT volumes can bias structural analysis and influence the outcome of longitudinal studies. On the other hand, presence of speckle noise that is characteristic of this imaging modality, leads to inaccuracies when traditional registration techniques are employed. Also, the lack of a well-defined ground truth makes supervised deep-learning techniques ill-posed to tackle the problem. In this paper, we tackle these issues by using deep reinforcement learning to correct inter-frame movements in an unsupervised manner. Specifically, we use dueling deep Q-network to train an artificial agent to find the optimal policy, i.e. a sequence of actions, that best improves the alignment by maximizing the sum of reward signals. Instead of relying on the ground-truth of transformation parameters to guide the rewarding system, for the first time, we use a combination of intensity based image similarity metrics. Further, to avoid the agent bias towards speckle noise, we ensure the agent can see retinal layers as part of the interacting environment. For quantitative evaluation, we simulate the eye movement artifacts by applying 2D rigid transformations on individual B-scans. The proposed model achieves an average of 0.985 and 0.914 for normalized mutual information and correlation coefficient, respectively. We also compare our model with elastix intensity based medical image registration approach, where significant improvement is achieved by our model for both noisy and denoised volumes.
Joint Segmentation and Uncertainty Visualization of Retinal Layers in Optical Coherence Tomography Images using Bayesian Deep Learning
Sep 12, 2018


Abstract:Optical coherence tomography (OCT) is commonly used to analyze retinal layers for assessment of ocular diseases. In this paper, we propose a method for retinal layer segmentation and quantification of uncertainty based on Bayesian deep learning. Our method not only performs end-to-end segmentation of retinal layers, but also gives the pixel wise uncertainty measure of the segmentation output. The generated uncertainty map can be used to identify erroneously segmented image regions which is useful in downstream analysis. We have validated our method on a dataset of 1487 images obtained from 15 subjects (OCT volumes) and compared it against the state-of-the-art segmentation algorithms that does not take uncertainty into account. The proposed uncertainty based segmentation method results in comparable or improved performance, and most importantly is more robust against noise.
Deep multiscale convolutional feature learning for weakly supervised localization of chest pathologies in X-ray images
Aug 22, 2018


Abstract:Localization of chest pathologies in chest X-ray images is a challenging task because of their varying sizes and appearances. We propose a novel weakly supervised method to localize chest pathologies using class aware deep multiscale feature learning. Our method leverages intermediate feature maps from CNN layers at different stages of a deep network during the training of a classification model using image level annotations of pathologies. During the training phase, a set of \emph{layer relevance weights} are learned for each pathology class and the CNN is optimized to perform pathology classification by convex combination of feature maps from both shallow and deep layers using the learned weights. During the test phase, to localize the predicted pathology, the multiscale attention map is obtained by convex combination of class activation maps from each stage using the \emph{layer relevance weights} learned during the training phase. We have validated our method using 112000 X-ray images and compared with the state-of-the-art localization methods. We experimentally demonstrate that the proposed weakly supervised method can improve the localization performance of small pathologies such as nodule and mass while giving comparable performance for bigger pathologies e.g., Cardiomegaly
Chest X-rays Classification: A Multi-Label and Fine-Grained Problem
Jul 24, 2018



Abstract:The widely used ChestX-ray14 dataset addresses an important medical image classification problem and has the following caveats: 1) many lung pathologies are visually similar, 2) a variant of diseases including lung cancer, tuberculosis, and pneumonia are present in a single scan, i.e. multiple labels and 3) The incidence of healthy images is much larger than diseased samples, creating imbalanced data. These properties are common in medical domain. Existing literature uses stateof- the-art DensetNet/Resnet models being transfer learned where output neurons of the networks are trained for individual diseases to cater for multiple diseases labels in each image. However, most of them don't consider relationship between multiple classes. In this work we have proposed a novel error function, Multi-label Softmax Loss (MSML), to specifically address the properties of multiple labels and imbalanced data. Moreover, we have designed deep network architecture based on fine-grained classification concept that incorporates MSML. We have evaluated our proposed method on various network backbones and showed consistent performance improvements of AUC-ROC scores on the ChestX-ray14 dataset. The proposed error function provides a new method to gain improved performance across wider medical datasets.
Elastic Registration of Medical Images With GANs
May 23, 2018



Abstract:Conventional approaches to image registration consist of time consuming iterative methods. Most current deep learning (DL) based registration methods extract deep features to use in an iterative setting. We propose an end-to-end DL method for registering multimodal images. Our approach uses generative adversarial networks (GANs) that eliminates the need for time consuming iterative methods, and directly generates the registered image with the deformation field. Appropriate constraints in the GAN cost function produce accurately registered images in less than a second. Experiments demonstrate their accuracy for multimodal retinal and cardiac MR image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge